Sauer:bis-Tris SDS-PAGE, the very best: Difference between revisions
No edit summary |
Wikified text |
||
| Line 1: | Line 1: | ||
Submitted by | Submitted by [[Sean Moore]] | ||
Based on work done by Tim Updyke and Sheldon Engelhorn for the Invitrogen Corporation (they bought Novex who developed it) and detailed in U.S. Patent 6,162,338. These gels are sold by Invitrogen as NuPAGE MES-Tris gels. | Based on work done by Tim Updyke and Sheldon Engelhorn for the [http://www.invitrogen.com/ Invitrogen Corporation] (they bought Novex who developed it) and detailed in U.S. Patent [http://patft.uspto.gov/netacgi/nph-Parser?Sect1=PTO1&Sect2=HITOFF&d=PALL&p=1&u=/netahtml/srchnum.htm&r=1&f=G&l=50&s1=6,162,338.WKU.&OS=PN/6,162,338&RS=PN/6,162,338 6,162,338]. These gels are similar to those sold by Invitrogen as NuPAGE MES-Tris gels. | ||
==Background== | |||
The pH of the separating gel in “standard” SDS-PAGE (a.k.a. Laemmli buffer system) is roughly 8-9 which is conducive to the deamination and alkylation of proteins, as well as reoxidation of reduced cysteines during electrophoresis. What this means is that your protein will form disulfide crosslinks during the stacking event because the protein migrates into the gel away from the reducing reagent in the sample buffer, and gets focused to a high concentration. Acrylamide gels cast in alkaline buffers are also unstable during long term storage, breaking down to acrylic acid after 1 to 2 months resulting in loss of pore size, poor resolution, and modified proteins. | The pH of the separating gel in “standard” SDS-PAGE (a.k.a. Laemmli buffer system) is roughly 8-9 which is conducive to the deamination and alkylation of proteins, as well as reoxidation of reduced cysteines during electrophoresis. What this means is that your protein will form disulfide crosslinks during the stacking event because the protein migrates into the gel away from the reducing reagent in the sample buffer, and gets focused to a high concentration. Acrylamide gels cast in alkaline buffers are also unstable during long term storage, breaking down to acrylic acid after 1 to 2 months resulting in loss of pore size, poor resolution, and modified proteins. | ||
| Line 16: | Line 16: | ||
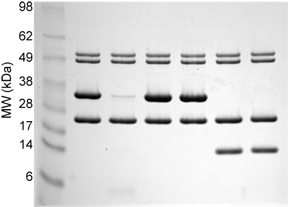
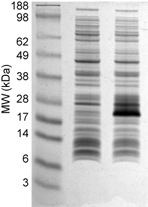
'''Sample bis-Tris MES-Tris Gels''' The first gel is of purified proteins. The second gel is of total ''E. coli'' lysate with and without an induced protein. Note the broad separation range of the molecular weight markers. | '''Sample bis-Tris MES-Tris Gels''' The first gel is of purified proteins. The second gel is of total ''E. coli'' lysate with and without an induced protein. Note the broad separation range of the molecular weight markers. | ||
==Materials== | |||
===acrylamide=== | |||
30% acrylamide | 30% acrylamide | ||
| Line 27: | Line 26: | ||
Alternately, you can use to 30/0.8 mix, just make the gel 12-15% final. | Alternately, you can use to 30/0.8 mix, just make the gel 12-15% final. | ||
===<u>5X</u> low-MW running buffer=== | |||
Use for separating small proteins 2-50 kDa. | Use for separating small proteins 2-50 kDa. | ||
| Line 35: | Line 34: | ||
0.5% SDS | 0.5% SDS | ||
No need to pH. | No need to pH. | ||
add sodium bisulf<u>ite</u> to 5 mM (add fresh before run)from a 1M stock. It's stinky. | add sodium bisulf<u>ite</u> to 5 mM (add fresh before run)from a 1M stock. It's stinky. | ||
===<u>5X</u> high-MW running buffer=== | |||
use for separating proteins >20 kDa. | use for separating proteins >20 kDa. | ||
| Line 50: | Line 48: | ||
add sodium bisulf<u>ite</u> to 5 mM (add fresh before run)from a 1M stock. It's stinky. | add sodium bisulf<u>ite</u> to 5 mM (add fresh before run)from a 1M stock. It's stinky. | ||
===200X running buffer reducing agent=== | |||
1 M sodium bisulfite | 1 M sodium bisulfite | ||
add to running buffer at 5 mM final concentration | add to running buffer at 5 mM final concentration | ||
===3.5X gel buffer=== | |||
1.25 M bis-Tris (pH 6.5-6.8 with HCl) | 1.25 M bis-Tris (pH 6.5-6.8 with HCl) | ||
| Line 62: | Line 59: | ||
note bis-Tris is Bis(2-hydroxyethyl) aminotris (hydroxymethyl) methane (e.g. Sigma catalog# B 7535). | note bis-Tris is Bis(2-hydroxyethyl) aminotris (hydroxymethyl) methane (e.g. Sigma catalog# B 7535). | ||
===sample buffer=== | |||
I routinely use the "standard" Laemmli 3X buffer. I think it does a better job reducing samples because of the higher pH. | I routinely use the "standard" Laemmli 3X buffer. I think it does a better job reducing samples because of the higher pH. | ||
==Casting and running gels== | |||
===Resolving:=== | |||
Mix: 1/3.5 vol. of 3.5X bis-Tris gel buffer, acrylamide to 8% (30:2.0) or 12-15% (30:0.8), and water to final volume. I make 3.75 mLs for each Bio-Rad Protean gel, and use 3.5 mLs per gel. | Mix: 1/3.5 vol. of 3.5X bis-Tris gel buffer, acrylamide to 8% (30:2.0) or 12-15% (30:0.8), and water to final volume. I make 3.75 mLs for each Bio-Rad Protean gel, and use 3.5 mLs per gel. | ||
| Line 83: | Line 80: | ||
Drain the butanol, rinse with water, wick the water from between the plates with a shred of filter paper. | Drain the butanol, rinse with water, wick the water from between the plates with a shred of filter paper. | ||
===Stacking:=== | |||
1X bis-Tris gel buffer, acrylamide solution to 4%, water. | 1X bis-Tris gel buffer, acrylamide solution to 4%, water. | ||
| Line 101: | Line 98: | ||
Rinse with water to remove unpolymerized acrylamaide. Remove comb. | Rinse with water to remove unpolymerized acrylamaide. Remove comb. | ||
===Running=== | |||
Fill both upper and lower buffer chambers with either MES-Tris or MOPS-Tris buffer. | Fill both upper and lower buffer chambers with either MES-Tris or MOPS-Tris buffer. | ||
Run at 150V constant. The Bromophenol blue runs around 3-5 kDa. | Run at 150V constant. The Bromophenol blue runs around 3-5 kDa. | ||
Revision as of 14:44, 26 August 2005
Submitted by Sean Moore
Based on work done by Tim Updyke and Sheldon Engelhorn for the Invitrogen Corporation (they bought Novex who developed it) and detailed in U.S. Patent 6,162,338. These gels are similar to those sold by Invitrogen as NuPAGE MES-Tris gels.
Background
The pH of the separating gel in “standard” SDS-PAGE (a.k.a. Laemmli buffer system) is roughly 8-9 which is conducive to the deamination and alkylation of proteins, as well as reoxidation of reduced cysteines during electrophoresis. What this means is that your protein will form disulfide crosslinks during the stacking event because the protein migrates into the gel away from the reducing reagent in the sample buffer, and gets focused to a high concentration. Acrylamide gels cast in alkaline buffers are also unstable during long term storage, breaking down to acrylic acid after 1 to 2 months resulting in loss of pore size, poor resolution, and modified proteins.
In this protocol, in-gel cysteine reoxidation is suppressed by casting and running under slightly acidic (~pH 6.5) conditions favoring cysteine protonation. Additionally, a reducing agent, sodium bisulfite, is included in the running buffer and will migrate into the gel and maintain a reducing environment. Another feature of this gel system is that the lower MW proteins near the buffer front do not accelerate towards the end of the run to the same degree as in Laemmli buffers. The result is higher resolution and a band distribution not unlike a gradient gel.
The Stacking and Resolving layers of the gel use the same buffer. This allows gels to be cast and stored for a long time (diffusion doesn't ruin the stacking chemistry). Also, the same tank running buffer is used at both the cathode and anode.
Sample bis-Tris MES-Tris Gels The first gel is of purified proteins. The second gel is of total E. coli lysate with and without an induced protein. Note the broad separation range of the molecular weight markers.
Materials
acrylamide
30% acrylamide 2% bis-acrylamide bis acrylamide can be added to preformulated acrylamide stocks (usually 30%:0.8% acrylamide/bis-acrylamide) to bring it up to the required amount.
Alternately, you can use to 30/0.8 mix, just make the gel 12-15% final.
5X low-MW running buffer
Use for separating small proteins 2-50 kDa.
250 mM MES 250 mM Tris 5 mM EDTA 0.5% SDS No need to pH.
add sodium bisulfite to 5 mM (add fresh before run)from a 1M stock. It's stinky.
5X high-MW running buffer
use for separating proteins >20 kDa.
250 mM MOPS 250 mM Tris 5 mM EDTA 0.5% SDS No Need to pH.
add sodium bisulfite to 5 mM (add fresh before run)from a 1M stock. It's stinky.
200X running buffer reducing agent
1 M sodium bisulfite add to running buffer at 5 mM final concentration
3.5X gel buffer
1.25 M bis-Tris (pH 6.5-6.8 with HCl)
note bis-Tris is Bis(2-hydroxyethyl) aminotris (hydroxymethyl) methane (e.g. Sigma catalog# B 7535).
sample buffer
I routinely use the "standard" Laemmli 3X buffer. I think it does a better job reducing samples because of the higher pH.
Casting and running gels
Resolving:
Mix: 1/3.5 vol. of 3.5X bis-Tris gel buffer, acrylamide to 8% (30:2.0) or 12-15% (30:0.8), and water to final volume. I make 3.75 mLs for each Bio-Rad Protean gel, and use 3.5 mLs per gel.
Add 25 μL of 10% APS per gel, mix in.
Add 7 μL TEMED
Pour gel and cover with ~1 mL of bis-Tris buffer saturated butanol.
Let set.
Drain the butanol, rinse with water, wick the water from between the plates with a shred of filter paper.
Stacking:
1X bis-Tris gel buffer, acrylamide solution to 4%, water.
Optional: Bromophenol Blue solution to make the stacking gel blue. It really helps when loading samples and doesn't affect the performance.
I make 2.5 mLs stacking per gel which is a bit too much.
Don't add APS until you are ready to start the polymerization. At the lower pH, the APS will start to polymerize the stacking gel while your resolving gel sets and will be all goopy.
Add 15-20 μL 10% APS, mix.
Add 7-10 μL TEMED, mix, pour onto the resolving layer, insert comb.
let set.
Rinse with water to remove unpolymerized acrylamaide. Remove comb.
Running
Fill both upper and lower buffer chambers with either MES-Tris or MOPS-Tris buffer.
Run at 150V constant. The Bromophenol blue runs around 3-5 kDa.