Griffin:siRNA transfection: Difference between revisions
| Line 130: | Line 130: | ||
==siRNA Controls== | ==siRNA Controls== | ||
===Mismatch or scrambled siRNAs=== | |||
These are often of somewhat limited value. A scrambled sequence is too unrelated to the 'active' probe to function as a truly informative control. Occasionally, a one- or two-base-pair change in the middle of the antisense can be a useful negative control if the siRNA effect is clearly ablated. However, such a change may have unanticipated effects by converting an siRNA into a miRNA (micro RNA) that inhibits translation through a pathway closely related to siRNA. | |||
===Basic controls=== | |||
siRNAs exert their effects through a growing number of surprisingly diverse mechanisms in addition to selective degradation of the targeted mRNA, such as specific effects at the chromatin level. Currently, siRNAs, unlike long double-stranded RNA (dsRNA), are not thought to trigger general translational attenuation through the interferon response. However, this also remains a hotly debated possibility for some short siRNAs, or at least mixed populations retaining some longer dsRNA species. In addition, the closely related miRNAs do function through target-specific translational attenuation. Thus, it is important to show reduction of expression at the mRNA and protein level, as well as a functional readout where available. If both message and protein are ablated, the response is 'classical' RNAi. In contrast, if only the protein is reduced, the chances are that a miRNA-related translational mechanism is at work. It should be noted that although this set of controls should be regarded as best practice, the functional control listed below renders analysis of the mRNA non-essential. Additional useful controls are available for unintentional activation of global translational repression through the interferon response (commercial assay kits or expression of unrelated proteins). Although centromeric or other emergent chromatin effects are harder to generate controls for, global gene expression may function as a control for any non-specific effects on gene expression. | |||
===Quantitative controls=== | |||
Titration of the siRNA is strongly encouraged. RNAi is often extremely effective already at minimal concentrations, and titration to the lowest possible levels reduces the chance of side effects as well as providing a graded readout of the effect. This is especially important because the RNAi machinery (the RISC complex in particular) is saturable, at least in some settings. Again, the rescue control outlined below is especially important when high levels of siRNA must be used. Protein levels should be assessed with quantitative techniques, such as quantitative western blotting, to allow for an accurate estimate of the level of reduction. | |||
===Functional controls=== | |||
The ultimate control for any RNAi experiment remains rescue by expression of the target-gene in a form refractory to siRNA (ideally within the physiological range). This can often be achieved by utilizing one or more silent third-codon point mutations within the targeted region, although controls for fortuitous miRNA effects are desirable. Translational effects can be avoided by utilizing siRNAs targeted against 3'-untranslated regions (UTRs), which are non-essential for rescue expression from a plasmid. The use of recently developed vector-based RNAi systems will alleviate some of the technical hurdles of rescue expression. | |||
===Multiplicity controls=== | |||
A good way to enhance confidence in RNAi data is to demonstrate a similar effect with two or more siRNAs targeted to different sites in the message under study. Alternatively, the RNAi approach is usefully supplemented by alternative methods, such as those described above. | |||
==siRNA related buffer and solutions== | ==siRNA related buffer and solutions== | ||
Revision as of 17:17, 23 October 2009
siRNA dependent translation initiation arrest is a user friendly technique that allows the scientist to determine the empirical moles of duplex/ number of cells, with a minimum number of steps toward achieving mRNA attenuation. shRNA transfection employs an ~10 kb transfer vector and additional antibiotic selection parameters that may influence off-targeting effects greater than conventional siRNA.
siRNA calculations
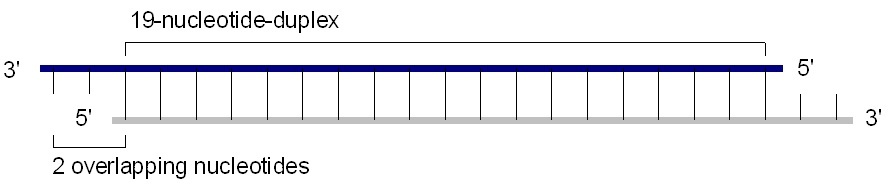
Chemically synthesized 21-25 nucleotide siRNA are suitable for transfection into dividing cells. A standard siRNA molecule is a chemically synthesized, 19 base pair double stranded RNA molecule with 2 base pair overhangs on the 3' ends (total of 21 base pair). Calculating the molecular weight of a single base pair based on chemical formula (at pH 7) gives ~615.4 Daltons/base pair on average+2 Na+ gives ~46 extra Daltons per base pair, or 661.4 Da/base pair total (average of GC & AT).
- Molarity of siRNA vialed: 10 uM ( uM/L )
- Volume after reconstitution: 330 uL
- Approximate mass of a mole of siRNA: 13800 g/mol ( 21nt X 660 g/base pair)
- Determine total mols per vial 10 um/L X 330 uL = 3.3 nm
- Convert mols to grams 3.3 nm X 13800 g/mol = 45.5 ug
- determine solution conc. 45.5 ug/ 330 uL = 0.138 ug/uL
After applying the right conversion factors (1Da = g/mol), we get 0.14 ug/uL of siRNA @ 10 uM.
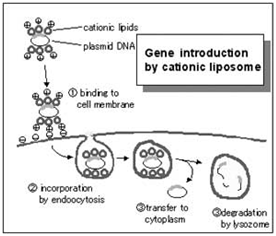
Cationic lipid-based transfection
Cationic lipid-based transfection reagents are suitable for transfecting into dividing cell cultures. Commercial examples include: Lipofectamine / L2000, Dharmafect, iFect, and TransIT TKO. All of these reagents are proprietary formulations that work by forming lipsomal vesicles that house the siRNA payload and bleb their way through the living cell membrane and into the cytoplasm. The efficiency of this process must be determined in order to have confidence in the knockdown effects. There are numerous commercial sources for transfection reagents for good reason; there are numerous cell types and lipsome structure will influence transfection efficiency in the multitude of experimental cell types that exist.
Verify transfection efficiency
The efficiency of transfection may depend on the cell type, but also on the passage number and the confluency of the cells. The time and the manner of formation of siRNA-liposome complexes (e.g. inversion versus vortexing) are also critical.
Verify the transfection efficiency with the use of a fluorescent dye-labeled siRNA; Control siRNA (FITC Conjugate)-A: sc-36869 (FITC, CY5, etc.). A flow cytometer can be used to measure cell transfection efficiency. The transfection efficiency must be measured 5-7 hours after cells are first exposed to the transfection reagent. This is important as the siRNA reagent complex with the siRNA will maximal enter the cell during this time frame. After 5-7 hours, the cells could begin to degrade or exotise the control siRNA-FITC conjugate and this will. When transfecting adherent cells, 5-7 hours after transfection, wash the slide then fix and mount the cells prior to visualization under a fluorescent scope.
1) In a six well tissue culture plate, seed 2e5 cells per well in 2 ml antibiotic-free normal growth medium supplemented with FBS.
2) Incubate the cells at 37° C in a CO2 incubator until the cells are 50 - 80% confluent. This will usually take 18-24 hours.
NOTE: Healthy and subconfluent cells are required for successful transfection experiments. It is recommended to ensure cell viability one day prior to transfection.
3) Prepare the following solutions:
Solution A: For each transfection, dilute 20-80 pmols siRNA into 100 µl siRNA Transfection Medium (serum/antibiotic-free medium). Solution B: For each transfection, dilute 2-8 µl of siRNA Transfection Reagent (brought to room temperature) into 100 µl siRNA Transfection Medium. Peak activity should be at about 6 µl siRNA Transfection Reagent.
4) Add the siRNA duplex solution (Solution A) directly to the dilute Transfection Reagent (Solution B) using a pipette. Mix gently by pipetting the solution up and down and incubate the mixture 15–45 minutes at room temperature.
5) Wash the cells once with 2 ml of serum/antibiotic-free medium and aspirate the medium and proceed immediately to the next step.
6) For each transfection, add 0.8 ml serum/antibiotic-free medium to each tube containing the siRNA Transfection Reagent mixture (Solution A + Solution B). MIx gently and overlay the mixture onto the washed cells. Total volume per well should be 1 mL.
NOTE: 20-80 pmols of siRNA in total volume of 1 ml (good for 1 well in a 6 well plate)= 20-80 nanomolar
7) Incubate the cells 5–7 hours at 37° C in a CO2 incubator.
8) Measure transfection efficiency: The transfection efficiency must be measured 5-7 hours after cells are first exposed to the transfection reagent.
Once the cell type is determined to be suitable for the chosen transfection reagent by measuring transfection efficiency, the RNAi procedure is ready to try.
siRNA transfection Procedure 6 well
Optimal conditions for transfection will vary. Amount of siRNA used for transfection may vary for each target protein and should be determined experimentally. Titrate the cell density parameter by trying different confluency of cells since different cell types can achieve an optimized knockdown at different cell densities. Cell density is an important variable. Different amounts of siRNA should be used (15, 30, 60, 90 nM) to determine what the minimal concentration necessary is to achieve efficient gene silencing. The amount of transfection reagent used per transfection is also an important variable and using more or less can improve transfection effiicency and cell viability.
1) In a six well tissue culture plate, seed 2 x 105 cells per well in 2 ml antibiotic-free normal growth medium supplemented with FBS.
2) Incubate the cells at 37° C in a CO2 incubator until the cells are 50 - 80% confluent. This will usually take 18-24 hours.
NOTE: Healthy and subconfluent cells are required for successful transfection experiments. It is recommended to ensure cell viability one day prior to transfection.
3) Prepare the following solutions:
Solution A: For each transfection, dilute 20-80 pmols siRNA into 100 µl siRNA Transfection Medium (serum/antibiotic-free medium). Solution B: For each transfection, dilute 2-8 µl of siRNA Transfection Reagent (brought to room temperature) into 100 µl siRNA Transfection Medium. Peak activity should be at about 6 µl siRNA Transfection Reagent.
4) Add the siRNA duplex solution (Solution A) directly to the dilute Transfection Reagent (Solution B) using a pipette. Mix gently by pipetting the solution up and down and incubate the mixture 15–45 minutes at room temperature.
5) Wash the cells once with 2 ml of serum/antibiotic-free medium and aspirate the medium and proceed immediately to the next step.
6) For each transfection, add 0.8 ml serum/antibiotic-free medium to each tube containing the siRNA Transfection Reagent mixture (Solution A + Solution B). MIx gently and overlay the mixture onto the washed cells. Total volume per well should be 1 mL.
NOTE: 20-80 pmols of siRNA in total volume of 1 ml (good for 1 well in a 6 well plate)= 20-80 nanomolar
7) Incubate the cells 5–7 hours at 37° C in a CO2 incubator.
8) Add 1 ml of normal growth medium containing 2 times the normal serum and antibiotics concentration (2x normal growth medium) without removing the transfection mixture. If toxicity is a problem, remove the transfection mixture and replace with 1x normal growth medium.
9) Incubate the cells for an additional 18–24 hours.
10) Aspirate the medium and replace with fresh 1x normal growth medium.
11) Assay the cells using the appropriate protocol 24–72 hours after the addition of fresh medium in the step above.
NOTE: The 12 well plate has a smaller well volume than a 6 well, so consider using less volume per well on a 12 well. siRNA concentration can affect cell viability. Experimental range that is common for siRNA is 1-100 nM. The issue of off-target effects must be addressed by running a control (non-targeting) siRNA. The control siRNA should be run in parallel at the same concentration as a way to ensure the result is authentic depending on what they are measuring (mRNA, protein, function).
Cells are incubated for 16 hours at 37°C and 5%CO2. Check for alteration in cell morphology or cytotoxic effects after siRNA transfection. This is a key step to visualize the cells and determine if they are healthy or if cytotoxicity is an issue. Cytotoxicity can be minimized by replacing the transfection solution with normal growth medium as early as 7 hours into transfection. In order to monitor gene silencing at the protein level proteins can be extracted after 24, 48 and 72 hours and measured by immunoblot. Quantitative RT-PCR is an alternative method to measure mRNA level.
siRNA transfection Procedure 24 well
- Seed adherent cells 1 day prior to transfection in 24-well plates using 500 µl of DMEM tissue culture medium supplemented with 10% FBS, without antibiotics.
- For one well of a 24-well plate, use 0.84 µg siRNA duplex (60 pmole in 3 µl annealing buffer).
- Mix 3 µl of 20 µM siRNA duplex (60 picomols) with 50 µl of Opti-MEM. In another tube, mix 3 µl of OLIGOFECTAMINE Reagent (or 3 to 3.5 µl Transit TKO) with 12 µl of Opti-MEM, incubate 7 to 10 min at room temperature.
- Combine the solutions and gently mix by inversion. Do not vortex. Incubate another 20 to 25 min at room temperature; the solution turns turbid.
- Then add 32 µl of fresh Opti-MEM to obtain a final solution volume of 100 µl. (The addition of 32 µl Opti-MEM is optional and serves only to adjust the total volume of cell culture medium to 600 µl after transfection.)
- Add the 100 µl of siRNA-OLIGOFECTAMINE to cultured cells (~40 to 50% confluent). Total volume is 600ul/well (60 pmol/600 ul =100 nM siRNA).
- After 5-7 hours, add normal growth medium containing normal serum and antibiotics concentration without removing the transfection mixture. If toxicity is a problem, remove the transfection mixture and replace with 1x normal growth medium.
- Incubate the cells for an additional 18–24 hours.
- Aspirate the medium and replace with fresh 1x normal growth medium.
- Assay the cells using the appropriate protocol 24–72 hours post transfection.
Working Cell Density Parameters
- Endothelial cells were seeded onto six well plates (2x10e5 cells per well) and incubated under normal growth conditions. The confluency of the transfected culture was about 80% at the time of transfection.
- Another condition reported to be successful was in a 12-well format at relatively low density 20-30% confluency of transformed mouse adherent cell; plated 4x10e4 cells per well 40 hours before transfection.
- Reverse transfection of adherent cells. Trypsinize cells and count the exact amounts per well, then perform the transfection as the cells are re-attaching to the plates. The advantage of this approach is knowing the cell count at the time of transfection and there may be a greater efficiency of uptake as the cells are re-attaching to the plates.
Analysis
Western blot and RT-PCR are good methods for knockdown analysis.
With regards to the kinetics of expression knockdown, it varies depending on the target protein. There are many factors that play a role on this. To name a few: target protein half life, constitutive protein expression versus transient protein expression, number of cells transfected (i.e., confluency), amount of siRNA used for transfection, transfection efficiency, siRNA sequence selection, etc. To simplify the process you can use a standard time of 48 hours post transfection to analyze knockdown.
Once knockdown is achieved, it will remain for as long as there is available siRNA inside the cells. Typically it will last for about 72 hours after transfection. For a longer knockdown, and for proteins with long half lives, a second or multiple siRNA transfections are recommended. If you do this, please make sure your control cells go through the same manipulation as your transfected cells, to subtract non-siRNA side effects.
When working on a protein that is critical for the cell, it is possible to observe unhealthy cells or high cell death after the siRNA transfection. In this case, it is recommended to check for knockdown sooner, or to decrease the amount of siRNA used to decrease the knockdown effect.
Eg-5 siRNA is a very useful control since it is critical for cell adhesion. For instance, validated expression knockdown 24 hours after transfection, which peaks at 48 hours, cells will remain viable, yet lose adherent phenotype. Eg-5 transfection causes loss of adherence of the cell.
siRNA Controls
Mismatch or scrambled siRNAs
These are often of somewhat limited value. A scrambled sequence is too unrelated to the 'active' probe to function as a truly informative control. Occasionally, a one- or two-base-pair change in the middle of the antisense can be a useful negative control if the siRNA effect is clearly ablated. However, such a change may have unanticipated effects by converting an siRNA into a miRNA (micro RNA) that inhibits translation through a pathway closely related to siRNA.
Basic controls
siRNAs exert their effects through a growing number of surprisingly diverse mechanisms in addition to selective degradation of the targeted mRNA, such as specific effects at the chromatin level. Currently, siRNAs, unlike long double-stranded RNA (dsRNA), are not thought to trigger general translational attenuation through the interferon response. However, this also remains a hotly debated possibility for some short siRNAs, or at least mixed populations retaining some longer dsRNA species. In addition, the closely related miRNAs do function through target-specific translational attenuation. Thus, it is important to show reduction of expression at the mRNA and protein level, as well as a functional readout where available. If both message and protein are ablated, the response is 'classical' RNAi. In contrast, if only the protein is reduced, the chances are that a miRNA-related translational mechanism is at work. It should be noted that although this set of controls should be regarded as best practice, the functional control listed below renders analysis of the mRNA non-essential. Additional useful controls are available for unintentional activation of global translational repression through the interferon response (commercial assay kits or expression of unrelated proteins). Although centromeric or other emergent chromatin effects are harder to generate controls for, global gene expression may function as a control for any non-specific effects on gene expression.
Quantitative controls
Titration of the siRNA is strongly encouraged. RNAi is often extremely effective already at minimal concentrations, and titration to the lowest possible levels reduces the chance of side effects as well as providing a graded readout of the effect. This is especially important because the RNAi machinery (the RISC complex in particular) is saturable, at least in some settings. Again, the rescue control outlined below is especially important when high levels of siRNA must be used. Protein levels should be assessed with quantitative techniques, such as quantitative western blotting, to allow for an accurate estimate of the level of reduction.
Functional controls
The ultimate control for any RNAi experiment remains rescue by expression of the target-gene in a form refractory to siRNA (ideally within the physiological range). This can often be achieved by utilizing one or more silent third-codon point mutations within the targeted region, although controls for fortuitous miRNA effects are desirable. Translational effects can be avoided by utilizing siRNAs targeted against 3'-untranslated regions (UTRs), which are non-essential for rescue expression from a plasmid. The use of recently developed vector-based RNAi systems will alleviate some of the technical hurdles of rescue expression.
Multiplicity controls
A good way to enhance confidence in RNAi data is to demonstrate a similar effect with two or more siRNAs targeted to different sites in the message under study. Alternatively, the RNAi approach is usefully supplemented by alternative methods, such as those described above.
siRNA related buffer and solutions
RNase-free H2O
- 0.1% DEPC in distilled H2O. Mix and leave at room temperature for 1 hour.
- Autoclave
- Cool to room temperature prior to use.
0.1% DEPC inactivates RNase contamination from common environmental sources and laboratory procedures. Autoclaving alone will inactivate a substantial amount of RNase as well.
Autoclaving inactivates DEPC by causing hydrolysis of diethylpyrocarbonate to yield CO2 and EtOH. DEPC has a half-life of ~30 minutes in water; 0.1% DEPC solution autoclaved for 15 minutes/liter can be assumed to be DEPC-free.
TE Buffer
10X TE Buffer (siRNA dilution buffer)
- 10 mM Tris, bring to pH 7.5 with HCl
- 1 mM EDTA
- RNase free water (DEPC water)
The EDTA in TE chelates Mg2+ and other divalent metals ions necessary for most causes of DNA and RNA degradation, suppressing these processes. Each EDTA molecule chelates one Mg2+ ion.
DNA is stored at pH 8 to reduce depurination, which is acid catalyzed, while RNA is stored at a slightly lower pH (7.5) because degradation of RNA is base-catalyzed. Most downstream reactions will not be influenced by the slightly different pH storage conditions.
References
Santa Cruz Biotechnology Protocols
Design References
siRNA Design Scoring
- Reynolds A, Leake D, Boese Q, Scaringe S, Marshall WS, and Khvorova A. Rational siRNA design for RNA interference. Nat Biotechnol. 2004 Mar;22(3):326-30. DOI:10.1038/nbt936 |
- Ui-Tei K, Naito Y, Takahashi F, Haraguchi T, Ohki-Hamazaki H, Juni A, Ueda R, and Saigo K. Guidelines for the selection of highly effective siRNA sequences for mammalian and chick RNA interference. Nucleic Acids Res. 2004;32(3):936-48. DOI:10.1093/nar/gkh247 |
- Amarzguioui M and Prydz H. An algorithm for selection of functional siRNA sequences. Biochem Biophys Res Commun. 2004 Apr 16;316(4):1050-8. DOI:10.1016/j.bbrc.2004.02.157 |
- Jagla B, Aulner N, Kelly PD, Song D, Volchuk A, Zatorski A, Shum D, Mayer T, De Angelis DA, Ouerfelli O, Rutishauser U, and Rothman JE. Sequence characteristics of functional siRNAs. RNA. 2005 Jun;11(6):864-72. DOI:10.1261/rna.7275905 |
- Kim DH, Behlke MA, Rose SD, Chang MS, Choi S, and Rossi JJ. Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat Biotechnol. 2005 Feb;23(2):222-6. DOI:10.1038/nbt1051 |
Immunostimulatory motifs
5'-UGUGU-3', 5'-GUCCUUCAA-3'
- Judge AD, Sood V, Shaw JR, Fang D, McClintock K, and MacLachlan I. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat Biotechnol. 2005 Apr;23(4):457-62. DOI:10.1038/nbt1081 |
- Hornung V, Guenthner-Biller M, Bourquin C, Ablasser A, Schlee M, Uematsu S, Noronha A, Manoharan M, Akira S, de Fougerolles A, Endres S, and Hartmann G. Sequence-specific potent induction of IFN-alpha by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat Med. 2005 Mar;11(3):263-70. DOI:10.1038/nm1191 |
Seed region sequence complimentarity
- Birmingham A, Anderson EM, Reynolds A, Ilsley-Tyree D, Leake D, Fedorov Y, Baskerville S, Maksimova E, Robinson K, Karpilow J, Marshall WS, and Khvorova A. 3' UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat Methods. 2006 Mar;3(3):199-204. DOI:10.1038/nmeth854 |
- Jackson AL, Burchard J, Schelter J, Chau BN, Cleary M, Lim L, and Linsley PS. Widespread siRNA "off-target" transcript silencing mediated by seed region sequence complementarity. RNA. 2006 Jul;12(7):1179-87. DOI:10.1261/rna.25706 |
- Anderson EM, Birmingham A, Baskerville S, Reynolds A, Maksimova E, Leake D, Fedorov Y, Karpilow J, and Khvorova A. Experimental validation of the importance of seed complement frequency to siRNA specificity. RNA. 2008 May;14(5):853-61. DOI:10.1261/rna.704708 |
- Ui-Tei K, Naito Y, Nishi K, Juni A, and Saigo K. Thermodynamic stability and Watson-Crick base pairing in the seed duplex are major determinants of the efficiency of the siRNA-based off-target effect. Nucleic Acids Res. 2008 Dec;36(22):7100-9. DOI:10.1093/nar/gkn902 |
Off-target Effects
- Khan AA, Betel D, Miller ML, Sander C, Leslie CS, and Marks DS. Transfection of small RNAs globally perturbs gene regulation by endogenous microRNAs. Nat Biotechnol. 2009 Jun;27(6):549-55. DOI:10.1038/nbt.1543 |
- Kenworthy R, Lambert D, Yang F, Wang N, Chen Z, Zhu H, Zhu F, Liu C, Li K, and Tang H. Short-hairpin RNAs delivered by lentiviral vector transduction trigger RIG-I-mediated IFN activation. Nucleic Acids Res. 2009 Oct;37(19):6587-99. DOI:10.1093/nar/gkp714 |
- van Dongen S, Abreu-Goodger C, and Enright AJ. Detecting microRNA binding and siRNA off-target effects from expression data. Nat Methods. 2008 Dec;5(12):1023-5. DOI:10.1038/nmeth.1267 |
- Tschuch C, Schulz A, Pscherer A, Werft W, Benner A, Hotz-Wagenblatt A, Barrionuevo LS, Lichter P, and Mertens D. Off-target effects of siRNA specific for GFP. BMC Mol Biol. 2008 Jun 24;9:60. DOI:10.1186/1471-2199-9-60 |