Uploads by Carolyne
From OpenWetWare
Jump to navigationJump to search
This special page shows all uploaded files.
| Date | Name | Thumbnail | Size | Description |
|---|---|---|---|---|
| 17:53, 30 April 2020 | Ver 2 Y41H and Q42E non-human primate mutants.png (file) | 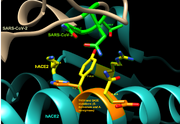 |
1.05 MB | This image shows how the alterations Y41H and Q42E (seen in S. bolivensis and A. nancymaae) affect the interactions that these primates' ACE2 may have with SARS-CoV-2 compared to human ACE2. |
| 17:32, 30 April 2020 | Ver 2 R357D and K353L non-human primate mutants.png (file) | 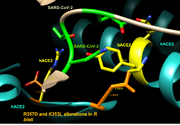 |
840 KB | This image shows how the alterations R357D and K353L (seen in R. bieti) affect the interactions that R. bieti ACE2 may have with SARS-CoV-2 compared to human ACE2. |
| 06:39, 23 April 2020 | C and N Terminus SC2.png (file) |  |
508 KB | Location of C and N terminus in SARS-CoV-2. |
| 06:36, 23 April 2020 | PredictProtein SARS-CoV-2 Predictions.png (file) | 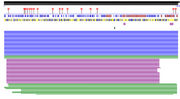 |
27 KB | Predictions for SARS-CoV-2 secondary structure based on amino acid sequence. |
| 06:25, 23 April 2020 | C and N Terminus for ACE2.png (file) |  |
216 KB | This image shows the location of the C and N terminus in ACE2. |
| 05:57, 23 April 2020 | Yan et al (2020) Figure 4A.png (file) | 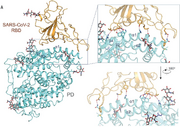 |
1.51 MB | Figure 4A from the Yan et. al. (2020) paper about SARS-CoV-2. This image shows the spike protein of SARS-CoV-2 interacting with ACE2 receptors. |
| 05:47, 23 April 2020 | Figure 4 recreation.png (file) | 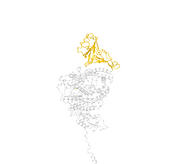 |
259 KB | This image recreates figure 4A from the Yan et. al. (2020) paper. |
| 12:41, 22 April 2020 | Week 13 UniProt Search Results.png (file) |  |
515 KB | This image shows the results generated after searching for "SARS-CoV" in UniProt |
| 11:45, 22 April 2020 | Week 13 ORF Image 2.png (file) |  |
142 KB | This image shows the results produced by the NCBI ORF Finder for SARS-CoV-2 spike protein. In this image, I have zoomed in on the ORF Viewer results. |
| 11:40, 22 April 2020 | Week 13 ORF Image 1.png (file) |  |
384 KB | This image shows the results produced by the NCBI ORF Finder for SARS-CoV-2 spike protein. |
| 07:57, 27 February 2020 | Egekeze On Nucleotide changes in V3 region (1).pptx.zip (file) | 1.31 MB | Presentation slides for week 7 | |
| 07:31, 27 February 2020 | Consensus Sequence Graph for Visits 1-4 at 17-32.png (file) |  |
16 KB | overall consensus for v1-4 |
| 07:31, 27 February 2020 | Nonprogressor Consensus Sequence Graph for Visits 1-4 at 17-32.png (file) |  |
16 KB | consensus for non progressors at v1-4 |
| 07:30, 27 February 2020 | Moderate Progressor Consensus Sequence Graph for Visits 1-4 at 17-32.png (file) |  |
15 KB | Consensus for mods at v1-4 |
| 07:29, 27 February 2020 | Rapid Progressor Consensus Sequence Graph for Visits 1-4 at 17-32.png (file) |  |
15 KB | Consensus for rapids at V1-4 |
| 06:39, 27 February 2020 | Colored CLUSTAL FORMAT alignment for visits 1 all sequences (1) (1).docx.zip (file) | 18 KB | Alignment for visit 1 sequences from subjects 2, 3, 5, 11, 13, and 14. Originally, I tried to highlight nucleotides at each non-starred position, but then I ended up analyzing the sequences in Excel so that I could highlight the cells easier. | |
| 06:36, 27 February 2020 | Hotspot 1 (position 17-32) from visits 1 and 4.zip (file) | 15 KB | Sequences of a hotspot (positions 17-32) in V3 sequence. Contains sequences from visit 1 and 4 for all subjects in project. | |
| 06:35, 27 February 2020 | Subject 1 Visits Sequence Alignment Analysis.xlsx.zip (file) | 52 KB | File containing alignments for clone sequences recovered during visit 1 for subjects 2, 3, 5, 11, 13, and 14. | |
| 06:30, 27 February 2020 | Phylo tree visit 1 AND 4 all sequences.png (file) | 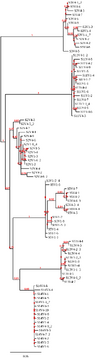 |
152 KB | The phylogenetic tree produced from all the sequences from subjects 2, 3, 5, 11, 13, and 14 at visit 1 and visit 4. |
| 06:30, 27 February 2020 | Phylo tree visit 4 all sequences.png (file) |  |
96 KB | The phylogenetic tree produced from all the sequences from subjects 2, 3, 5, 11, 13, and 14 at visit 4. |
| 06:29, 27 February 2020 | Phylo tree visit 1 all sequences.png (file) |  |
61 KB | The phylogenetic tree produced from all the sequences from subjects 2, 3, 5, 11, 13, and 14 at visit 1. |
| 21:33, 19 February 2020 | Subject 12 phylo tree all visits .png (file) |  |
19 KB | This phylogenetic tree shows the evolutionary relationships between all of the clones taken from Subject 12 over the course of the study. |
| 21:31, 19 February 2020 | Subject 11 Phylogenetic tree.png (file) |  |
16 KB | This phylogenetic tree displays the evolutionary relationships between all of the clones taken from Subject 11 over the course of the study. |
| 21:28, 19 February 2020 | Subject 6 phylo tree all visits.png (file) |  |
28 KB | This phylogenetic tree displays the evolutionary relationships between all clones found in Subject 6 over the course of the study. |
| 23:17, 13 February 2020 | Markham SMultiV1C1-3.png (file) | 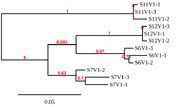 |
9 KB | Phylogenetic tree generated from analyzing sequences of clones 1-3 from Subjects 6, 7, 11, and 12. Clone sequences were recovered from subject samples obtained during visit 1. |
| 22:41, 13 February 2020 | Markham S1V1C2-7 Phylo Tree.png (file) | 6 KB | The phylogenetic tree produced after aligning the HIV clone sequences 2-7 from Subject 1. Clone sequences were obtained from samples taken during the subject's first visit. | |
| 05:25, 23 January 2020 | Egekeze Working CV.pdf (file) | 67 KB | Here is a draft of my CV, updated on January 22, 2019. | |
| 04:05, 23 January 2020 | Egekeze OWW Photo copy.jpg (file) |  |
1.31 MB | Reverted to version as of 19:54, 22 January 2020 (PST) |