Sosnick:Research
| |
|
|
|
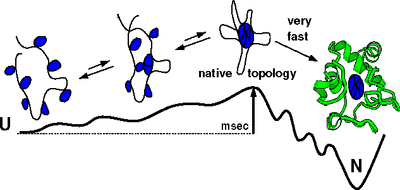
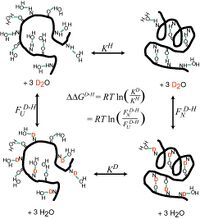
In the past 7 years, we have learned to direct folding and dynamics of these biopolymers. We have developed Ψ-analysis , a widely applicable method that identifies structures along folding routes. The method utilizes designed metal binding sites in conjunction with a new theory to analyze the accompanying non-linear Brønsted behavior. It has provided one of the most accurate descriptions of a protein folding pathway yet obtained. In RNA folding, we generated atomic level models of folding intermediates and revealed the principles of tertiary RNA stability using homologous RNAs from mesophilic and thermophilic organisms. Our studies have led to the concept of thermodynamic and kinetic hotspots. These fundamental studies have helped us examine how non-coding RNAs may fold in the cell. We found that species-specific pausing during RNA transcription improves the folding behavior, thereby suggesting that RNA sequences have co-evolved with the pausing properties of their cognate RNA polymerases. In addition to continuing the experimental protein and RNA folding studies, new research areas in the Sosnick lab involve the rational design of proteins and RNAs to control their structure and function, and the expansion of the computational component. We have developed a method to “fuse” two proteins together in order to create a single light-triggered allosteric repressor protein. We are also predicting protein structures by simulating folding pathways, taking advantage of our experimental results and an approach that dynamically integrates local and global information. In addition, we are refining crystal structures and homology models using our new move-set and statistical potentials. With Wah Chiu (Baylor), we recently have pushed the molecular weight limit for cryoEM studies down by 5-fold. This work opens up a whole new avenue of structural studies on non-coding RNAs. Our long-term goal is to create residue-level structural models using the cryoEM images, sequence information, and all-atom simulations, building on our modeling studies of RNA folding intermediates. Graduate students in my lab enter through the new Biophysical Sciences Graduate Program, in addition to the Programs in Biochemistry & Molecular Biology, Chemistry, and Physics. | |
|---|---|
| Do a PubMed search for "Sosnick TR." | |
| Home | Research | People | Internal | Contact |