Sbb14 Neeka Meshgin
Tuesday Feb. 11
Searched web for thermophilic bacteria.
Options:
Borrelia burgdorferi
Aquifex aeolicus
Thermus thermophilus
Thermatoga maritima
Contacted Professor Wemmer to see if he has access to aquifex since we can't find it on atcg.
Thursday Feb. 13
Chose thermus thermophilus & thermatoga maritima. Split up 20 amino acids and 2 bacteria between 4 teams.
Friday Feb. 14
Split up 10 tRNA synthetases between group.
adrianna:M N neeka: P W daniel: R S kevin: T V akash:Q Y
Did Pubmed search for DNA sequences for prolyl and tryptophanyl tRNA synthetases of thermus thermophilus. Found them. No internal restriction sites for Nco1 and EcoR1.
Neeka Meshgin 13:38, 18 February 2014 (EST)
Designed oligos.
Forward prolyl tRNA synthetase
ccaaaCCATGGatggcgaaggagaagggcct
Reverse prolyl tRNA synthetase gctagGAATTCtcagtacgccttggcgaaga
Forward tryptophanyl tRNA synthetase
ccaaaCCATGGatgaagcgggtgctttccgg
Reverse tryptophanyl tRNA synthetase gctagGAATTCtcaccggaggaccggccgct
Neeka Meshgin 13:44, 20 February 2014 (EST)
Finished designing oligos and writing construction files. Edited my oligos for tryptophanyl tRNA synthetase to remove internal restriction site. Double-checked each other's oligos.
Construction file for tryptophanyl tRNA synthetase:
PCR TTHA1227F/ TTHA1227internalR on TTHA1227 ( 974 bp, A) PCR TTHA1227internalF/TTHA1227R on TTHA1227 (83 bp, B) PCR TTHA1227F/ TTHA1227R on A+B (1036 bp, pcrpdt) Digest prcpdt (Nco1/ EcoR1, L, 1005 pcrdig) Digest pBAD:myc-HisA (Nco1/EcoR1, 3736+317, plasdig) Ligate pcrdig and plasdig, product is TTHA1227-pBAD:myc-HisA
>TTHA1227F forward oligo
ccaaaCCATGGatgaagcgggtgctttccgg
>TTHA1227R reverse oligo
gctagGAATTCtcaccggaggaccggccgct
>TTHA1227internalF forward removing Nco1 site
caggcgacGatggaggaggtg
>TTHA1227internalR reverse removing Nco1 site
cacctcctccatCgtcgcctg
>TTHA1227 tryptophanyl tRNA synthetase of thermus thermophilus
>pBAD:myc-HisA
Construction file for prolyl tRNA synthetase
PCR TTHA0115-F/ TTHA0115-R (1456 bp, pcrpdt) Digest pcrpdt (Nco1/EcoRI, 1425+25+6, L, pcrdig) Digest pBAD:myc-HisA (Nco1/EcoRI, 3736+317, L, vectdig) Ligate pcrdig and vectdig (TTHA0115-pBAD:myc-HisA)
>TTHA0115F forward oligo
ccaaaCCATGGatggcgaaggagaagggcct
>TTHA0115R reverse oligo
gctagGAATTCtcagtacgccttggcgaaga
>TTHA0115 prolyl tRNA synthetase of thermus thermophilus
>pBAD:myc-HisA
Thursday Feb. 27
Watched demo of mini prep and gel purification.
Did mini prep protocol.
Tuesday March 4
Set up PCR. First made a "master mix" of ddH2O, buffer, and dNTP's for our group. Then diluted my oligos to 10 μM. Then combined "master mix," oligos, template DNA, and enzyme in a PCR reaction tube. Then placed the reaction tube in the thermocycler.
Thursday March 6
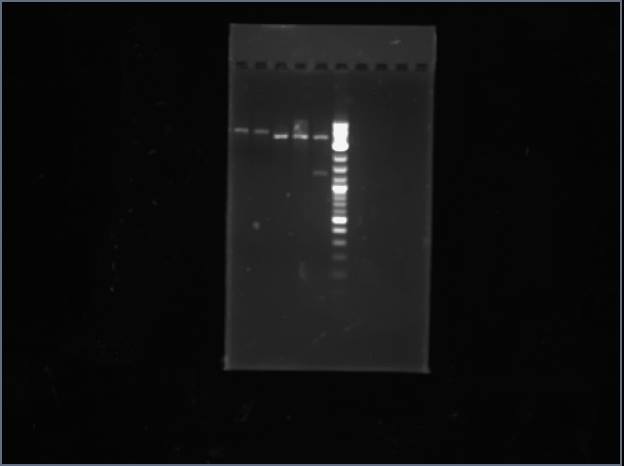
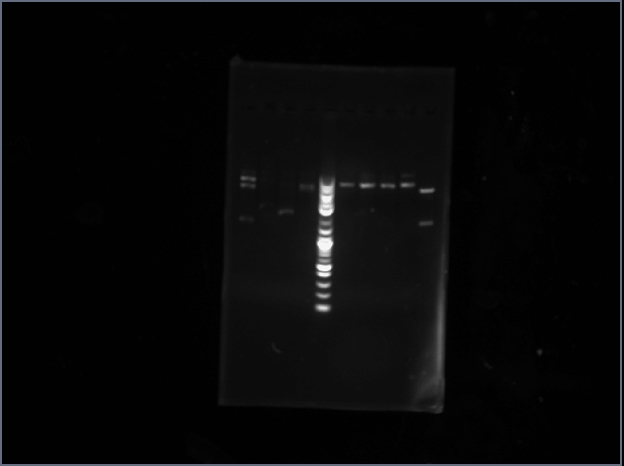
Ran a gel for our PCR products from Thursday. Out of my tRNA synthases, tryptophan worked, proline did not (lane 1).
Friday March 7
Re-did PCR for proline. Did Zymo clean-up of my successful tryptophan.
Tuesday March 11
Did zymo cleanup of proline PCR product.
Digested tryptophan, and did zymo cleanup of digested tryptophan.
Thursday March 13
Ran a gel for proline PCR product. Gel displayed multiple products, ranging from 1400-2000 bp (should only have 1400).
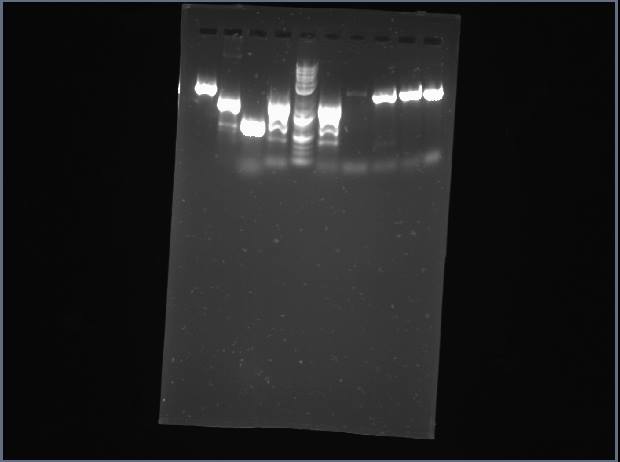
Proline is shown above in lanes 4 and 6.
Re-did PCR for proline, (same protocol, except using 1 μL of DMSO and subtracting 1 μL of water) to prevent possibility that oligo's are annealing upstream of target sequence.
Tuesday March 18
Ran a gel for proline PCR product. 2 distinct bands were shown, and I cut out the 1400 bp band. Did zymo cleanup of proline PCR product from the gel.
Thursday March 20
Digested proline PCR product, and then did zymo cleanup of proline digest.
Ligated typtophan digest with vector digest following ligation protocol.
Followed transformation protocol. Also did a negative control (just vector, no ligated tRNA synth gene.) Streaked plasmids onto agar plates. Incubated at 37°C overnight.
Friday March 21
Checked plates--none have colonies.
Tuesday April 1
Ligated proline digest with vector digest following ligation protocol. Followed transformation protocol for both proline and tryptophan. Streaked transformed cells on agar plates and incubated overnight.
Thursday April 3
Colonies successfully grew on plates. Picked out round, well-isolated colonies to do miniprep later. Picked out 4 colonies for proline and 4 for tryptophan, and suspended each colony in 4 mL of LB media and ampicillin. Incubated overnight at 37°C.
Tuesday April 8
Colonies successfully grew in each test tube. Re-suspended cultures, and did miniprep purification of DNA following protocol.
Thursday April 10
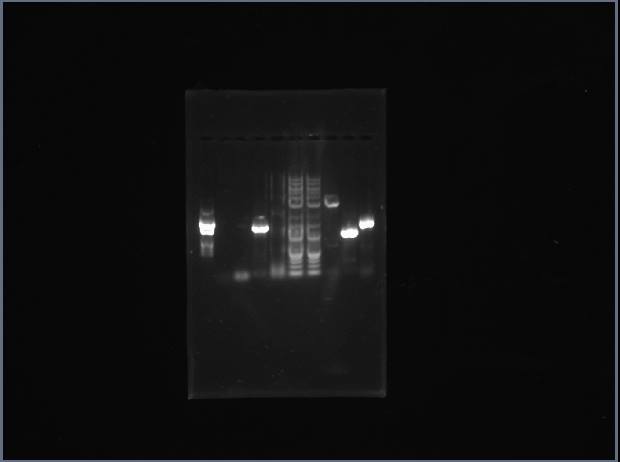
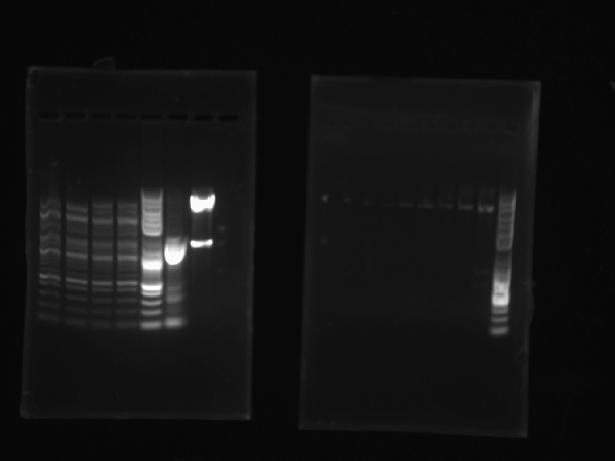
Ran an analytical gel to check if cells were successfully transformed. Digested miniprep DNA samples (Trp #2, Trp $3, Pro #2, Pro #4) and vector DNA with NcoI/EcoRI using digestion protocol. Expecting to see 1400 bp band for proline trna synthetase gene and 1000 bp band for tryptophan. Gel shows unusual results--several small bands in each lane, resembling the ladder. Will re-do analytical gel.
From left: Trp #2, Trp $3, Pro #2, Pro #4. The rest belong to other group members.
Tuesday April 15
Since last gel didn't work, did another miniprep purification of DNA using protocol for all samples (Pro#1-4 and Trp#1-4)
Digested purified DNA samples with NcoI/EcoRI.
Thursday April 17
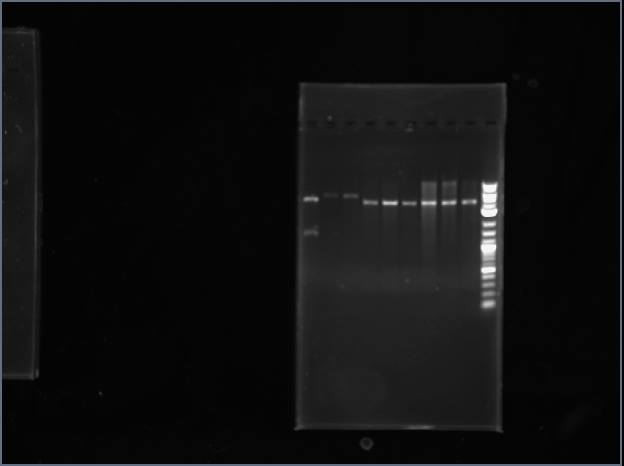
Ran a gel for digested samples.
Left to right: Pro#1, 2, 3, 4, ladder, Trp #1, 2, 3, 4, digested vector
Tuesday April 22
Digested miniprep samples Trp#1-4 using NcoI/EcoRI.
Turned in my predicted sequence for Pro#1 to get sequenced.
Thursday April 24
Ran gel of digested products, Trp #1-4.
From left: vector digest, Trp#1, Trp #2. Lanes 4-9 are other group member's samples. Lane 10 is the ladder.
From left: Trp#3, Trp#4, Lanes 3-4 are other group member's samples. Lane 5 is vector dig. Lane 6 is ladder.