Sbb14-Kelvin Li
Organism
- Thermotoga maritima MSB8
- ATCC number: 43589D-2
- PubMed accession number (Nucleotide database): AE000512
- genome (Genbank format): File:Thermotoga maritima MSB8.gb
Translation table
Bacterial, Archaeal and Plant Plastid Code
AAs = FFLLSSSSYY**CC*WLLLLPPPPHHQQRRRRIIIMTTTTNNKKSSRRVVVVAAAADDEEGGGG Starts = ---M---------------M------------MMMM---------------M------------ Base1 = TTTTTTTTTTTTTTTTCCCCCCCCCCCCCCCCAAAAAAAAAAAAAAAAGGGGGGGGGGGGGGGG Base2 = TTTTCCCCAAAAGGGGTTTTCCCCAAAAGGGGTTTTCCCCAAAAGGGGTTTTCCCCAAAAGGGG Base3 = TCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAG
valyl-tRNA synthetase
- Protein Sequence:
MAELSTRYNPAEIETKWYRYWEEKGYFTPKGVGEKFSIVIPPPNITGRIHMGHALNITLQDIVVRYKRMK GYDVLWVPGEDHAGIATQNAVEKFLLQTQGKTREEIGREKFLEITWEWANKYRREIREQIKALGASVDWT RERFTLDEGLSRAVRKVFVELYRKGLIYRGKYIVNWCPRCKTVLSDEEVEHKEHKSKLYYVKYPVKDSDE YIVVATTRPETMLGDTAVAVHPEDERYKNFVGKTLILPLVGREIPVVADKYVDPKFGTGAVKVTPAHDPN DYLIAQRHNLPMIEIFDDNARINENGGKYKGLDRYEAREKIVKDLEEQGFLVKIEDYTHSVGHCYRCDTV IEPKLSDQWFVSTKPLAKRAIEAVENGEIRFFPERWTKVYLNWMYEIRDWCISRQLWWGHRIPVWYCQDC GHLNVSEEDVEKCEKCGSTNLKQDEDVLDTWFSSALWPFSTLGWPEETEDLKRYYPTDLLVTGFDIIFFW VARMIMMGYEFMNDKPFSHVYIHQLVRDKYGRKMSKSLGNGIDPLEVIDEYGADPMRFTLAILAAQGRDI KLDPRYFDAYKKFANKIWNATRFVLMNLEDYKEVPLENLKTVDKWILTRLNKTVEEVTNALENYDFNIAA RTIYNFFWDDFCDWYIEASKPRLKTEERNLVQTVLVKVLDASLRLLHPFMPFLTEELWQKLPVAGESITI AKWPEIERELIDETAEKEFTRLMNMVRGVRNVRAEMNLPQSQRVKVYIKGYEVTEEEELLLKTLGNIEEV SFVNEKPPKTATAYVEEEIEAYVDLGGLIDFEKEKERLKQIMEKIQKEIDRLEKKLANKDFVEKAPEEVV EETKEKLNTNRERLARLESILRDLE
threonyl-tRNA synthetase
- Protein Sequence:
MKIKVKLPDGKEKEYDRGITPAEIAKELGIKKAIGAVVNGELWDLKRPIENDCELRLVTLEDPEAPEFYR HTMAHILAQAVMRLYGKENVKLGIGPTIENGFYYDFDIKNGRLTEEDLPKIEQEMKKIIKENLPIERKEI SKEEARELFRDQPYKLELIEEIEGNRVTIYRQGEFVDLCRGPHLPSTGIVKHFKLLSVSGAYWRGSEKNP MLTRVYGTAFAKKEDLDNYLKFLEEAQRRDHRKLGPQLELFMLNTEYAPGMPFFLPKGVVVLNELMKFSR ELHRERGYQEIFTPLIMNEQLWKISGHWDHYAENMYFIEKDEERYAVKPMNCPGHILVYKSRTVSYRDLP LRFFEFGRVHRYERSGVLHGLMRVRSFTQDDAHIFCTPDQIEEEILGVLDLINTIYSQFGFTYRVELSTM PEDHMGDEAIWEKATTALKNALERAGLSYKVNEGEGAFYGPKIDFHIRDSIGREWQCATIQLDFMMPEKF NVTYIGPDNKEHRAVMIHRAIYGSLERFFGILIEHFAGAFPTWLAPIQVAVIPISEKHNDGAEKVARRIS QEGFRVFFDNRRETLGYRIRQAQTQKIPYMIIIGDKELESGKISVRTRTGKEIKDVDPEHFVETLRNEVL SRKLELSMEG
Construction files
Plasmid: pBAD OLED (a compatible derivative of pBAD/myc-His A)
Two references for pBAD/myc-His_A. [1] [2]
Construction of threonyl-tRNA synthetase part
SOE'ing of one NcoI internal restriction site.
PCR externF/internR on genome (241 bp, A) PCR internF/externR on genome (1731 bp, B) PCR externF/externR on A + B (1944 bp, pcrproduct) Digest pcrproduct (NcoI/EcoRI, L, pcrdigest) Digest pBAD (NcoI/EcoRI, 4053+41, L, plasmiddigest) Ligate pcrdigest and plasmiddigest (5981 bp, finalplasmid) --------------------------------------------- >externF ThrRS_NcoI_Fwd ccagtccatggctaagataaaggtgaagcttccag >externR ThrRS_EcoRI_Rv ccagtgaattcttatccctccattgaaagttcg >internF ThrRS_SOE-NcoI_Fwd ctacagacacacGatggcacacatcctc >internR ThrRS_SOE-NcoI_Rv gaggatgtgtgccatCgtgtgtctgtag >pBAD pBAD myc-His A >genome thermotoga maritima MSB8 genome
Construction of valyl-tRNA synthetase part
SOE'ing of one EcoRI internal restriction site.
PCR externF/internR on genome (2176 bp, A) PCR internF/externR on genome (468 bp, B) PCR externF/externR on A + B (2161 bp, pcrproduct) Digest pcrproduct (NcoI/EcoRI, L, pcrdigest) Digest pBAD (NcoI/EcoRI, 4053+41, L, plasmiddigest) Ligate pcrdigest and plasmiddigest (6653 bp, finalplasmid) --------------------------------------------- >externF ValRS_NcoI_Fwd ccagtccatggcagaactctcgacgagatac >externR ValRS_EcoRI_Rv ccagtgaattctcattctagatctctgaggatg >internF ValRS_SOE-EcoRI_Fwd cgcggagaaggaGttcaccaggctcatg >internR ValRS_SOE-EcoRI_Rv catgagcctggtgaaCtccttctccgcg >pBAD pBAD myc-His A >genome thermotoga maritima MSB8 genome
2014-02-27
- Miniprepped parent plasmid from E. coli using protocol at Template:SBB-Protocols_Micro3.
- Tube label:
KL 4, KL pBad OLED grp 4
- Tube label:
- Stored in freezer.
2014-03-04
- Received dry oligos.
- Spun tubes for approximately 30 seconds.
- Made stock solutions. Added water according to the table below, such that the concentration of each stock soution is 100μM.
| Label | Quantity (nanomoles) | ddH2O added |
ThrRS_NcoI_Fwd |
28.5nmol | 285μL |
ThrRS_EcoRI_Rv |
30.9nmol | 309μL |
ThrRS_SOE-NcoI_Fwd |
33.9nmol | 339μL |
ThrRS_SOE-NcoI_Rv |
38.5nmol | 385μL |
ValRS_NcoI_Fwd |
35.8nmol | 358μL |
ValRS_EcoRI_Rv |
32.7nmol | 327μL |
ValRS_SOE-EcoRI_Fwd |
29.0nmol | 290μL |
ValRS_SOE-EcoRI_Rv |
33.5nmol | 335μL |
- Vortexed and spun tubes.
- For each stock solution, created dilutions to 10μM consisting of:
- 1μL oligo stock
- 10μL ddH2O
- Setup expand PCRs according to protocol in "PCR in Practice -> Basic PCR for cloning". Labeled tubes:
| PCR tube label | Oligo 1 | Oligo 2 |
T A, 4KL
|
ThrRS_NcoI_Fwd
|
ThrRS_SOE-NcoI_Rv
|
T B, 4KL
|
ThrRS_SOE-NcoI_Fwd
|
ThrRS_EcoRI_Rv
|
V A, 4KL
|
ValRS_NcoI_Fwd
|
ValRS_SOE-EcoRI_Rv
|
V B, 4KL
|
ValRS_SOE-EcoRI_Fwd
|
ValRS_EcoRI_Rv
|
- Stored oligo stock and PCR setups in freezer.
2014-03-06
Thawed and ran PCRs created on 2014-03-04. Used 2K55 program.
2014-03-07
The original SOE'ing PCR protocol is at Template:SBB-Protocols_PCR3.
- Retrieved finished PCR products (total of four tubes) from 2014-03-06.
- Spun tubes for approximately 15 seconds.
- Setup 1% agarose gel with 10 wells and 1x TAE buffer solution.
- For each of the four PCR products, mixed the following in a new tube:
- 1μL 6x loading dye
- 6μL PCR product
- Stored original PCR product tubes in freezer.
- Spun new, PCR-dye mix tubes for approximately 10 seconds.
- Loaded one gel well for each PCR-dye mix.
- Started gel at approximately 13:15. Ended at approximately 13:40.
- Gel ran at 150 volts.
- Visualized gel with long blue light (called "trans UV" on machine)
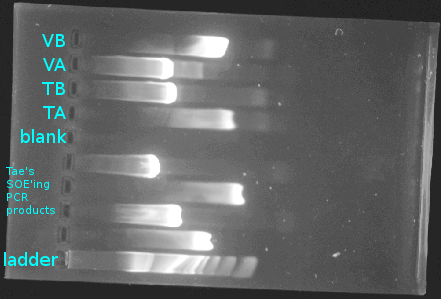
- Cut out my four bands and put into two 1.5mL tubes, labelled as follows:
| combined 1.5mL tube label | PCR product 1 original tube label | PCR product 2 original tube label |
VA+VB, 4KL |
V A, 4KL |
V B, 4KL
|
TATB, 4KL |
T A, 4KL |
T B, 4KL
|
Note that the two tubes each contain two distinct PCR products.
- Added 650μL ADB buffer to each tube.
- Melted bands by partially submerging tubes in a warm water bath.
- Stored the two tubes in freezer. These are the templates for the third (and last) SOE'ing PCR step.
2014-03-11
- Retrieved two tubes from freezer from 2014-03-07:
VA+VB, 4KLandTATB, 4KL. - Finished remainder of Zymo gel purification: Arking:JCAProtocols
- Skipped adding of 250μL isopropanol, even though the tube
TATB, 4KLcontains DNA that is less than 300bp. - Tranferred to Zymo column, in collection tube.
- Spun at full speed for one minute. Discarded waste.
- Added 200μL Zymo wash buffer
- Spun at full speed for 30 seconds. Discarded waste.
- Again added 200μL Zymo wash buffer.
- Again spun at full speed for 30 seconds. Discarded waste.
- Spun at full speed for 2 minutes to dry.
- Eluted DNA with 50μL ddH2O into fresh Eppendorf tubes: 1 minute full speed spin.
- Skipped adding of 250μL isopropanol, even though the tube
- Retrieved oligo stock from freezer; created two 10μM dilutions with two oligos in each:
| Mixed dilution tube label | Stock Oligo 1 | Stock Oligo 2 | ddH2O |
T oligo dilute |
ThrRS_NcoI_Fwd (1μL) |
ThrRS_EcoRI_Rv (1μL) |
18μL |
V oligo dilute |
ValRS_NcoI_Fwd (1μL) |
ValRS_EcoRI_Rv (1μL) |
18μL |
- Setup PCR reactions:
| PCR tube label | Mixed oligo dilution tube | Template tube | 10x Expand buffer ("2") | 2mM dNTP's | Expand polymerase ("1") |
T_PCR, 4KL |
T oligo dilute (2μL) |
TATB, 4KL (24μL) |
3.3μL | 3.3μL | 0.5μL |
V_PCR, 4KL |
V oligo dilute (2μL) |
VA+VB, 4KL (24μL) |
3.3μL | 3.3μL | 0.5μL |
- Ran on 4K55 program.
- Discarded mixed oligo dilutions (
T oligo diluteandV oligo dilute). - After PCR completes, the products in tubes
T_PCR, 4KLandV_PCR, 4KLcorrespond topcrproductin construction files.
2014-03-13
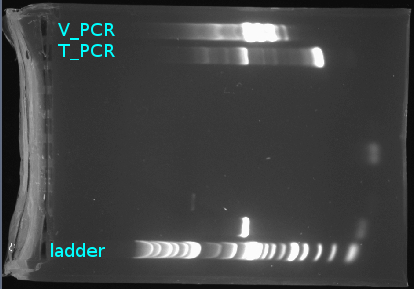
- Retrieved the two PCR products from 2014-03-11:
T_PCR, 4KLandV_PCR, 4KL - Ran analytical gel at 160 volts in the two right-most lanes:
| DNA | Lane | ddH2O | 6x loading dye |
T_PCR, 4KL (2μL) |
second from right-most | 6μL | 1μL |
V_PCR, 4KL (2μL) |
right-most | 6μL | 1μL |
Started at 10:55 AM and ended 11:47 AM.
- Visualized gel under long blue:
- Stored tubes in freezer.
2014-03-18
- Performed regular Zymo cleanup of tubes
T_PCR, 4KLandV_PCR, 4KL.- Used 180μL ADB buffer and eluted with 33μL ddH2O.
- Setup 30μL digestion.
- Placed both tubes on ice.
- In each tube, added:
| 15μL | DNA |
| 3μL | 10x NEB buffer "2" |
| 1.5μL | EcoRI |
| 1.5μL | NcoI |
| 9μL | ddH2O |
- Started digestion at 11:00 AM. Ended 12:00 PM. Held at 37°C throughout.
- Performed Zymo cleanup with 30μL ddH2O elution.
- Labeled tubes:
T dig, 4KLandV dig, 4KL
- Labeled tubes:
- Stored tubes in freezer.
2014-03-20
- Made 10-well 1% agarose gel with:
| 1 gram | agarose powder |
| 100mL | 1x TAE buffer |
| 10μL | GelGreen dye |
- Retrieved tubes
T dig, 4KLandV dig, 4KLfrom freezer. - Prepared tubes for loading into gel:
- 30μL digestion product
- 3μL 6x loading dye
- Loaded 3 wells for each tube (6 wells total). Each well was loaded with 10μL of contents.
- Ran gel at 155 volts
- Started 11:00 AM
- Stopped 11:48 AM
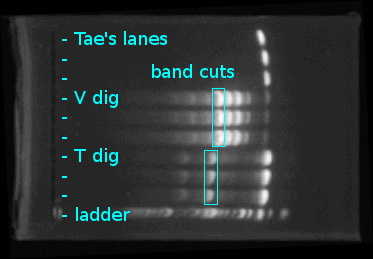
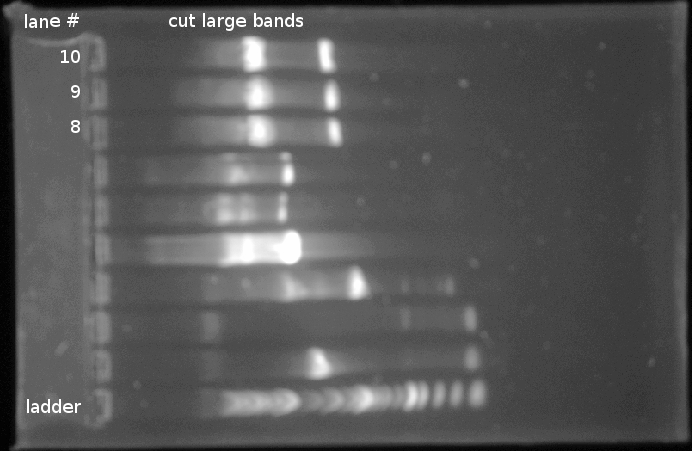
- Cut bands as shown below:
- Sorted bands into two tubes (one for each DNA)
- Melted bands in 650μL ADB buffer at 51°C
- Performed regular zymo cleanup
- Eluted each DNA into 15μL ddH2O
- Stored tubes in freezer
| Cleaned-up digest tube label | Original digest tube label |
T pure, 4KL |
T dig, 4KL
|
V pure, 4KL |
V dig, 4KL
|
2014-04-01
- Retrieved mini-prepped vector plasmid from freezer.
- tube label:
KL 4, KL pBad OLED grp 4
- tube label:
- Prepared two identical 30μL digests (referred to as "1" and "2") on ice:
- 15μL plasmid
- 3μL 10x NEB Buffer "2"
- 9μL ddH2O
- 1.5μL EcoRI
- 1.5μL NcoI
- Incubated at 37°C
- Started digests "1" and "2" at 10:17 AM and 10:31 AM, respectively.
- Stopped both at 11:50 AM
- Made 8-well gel
- 0.5g agarose powder
- 50mL 1x TAE buffer
- 5μL GelGreen dye
- Added 3μL 6x gel loading dye to each completed digest tube.
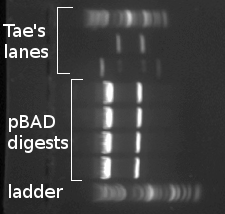
| Lane | Contents |
| 1 | 5μL ladder |
| 2, 3 | 15μL per well, digest "2" |
| 4, 5 | 15μL per well, digest "1" |
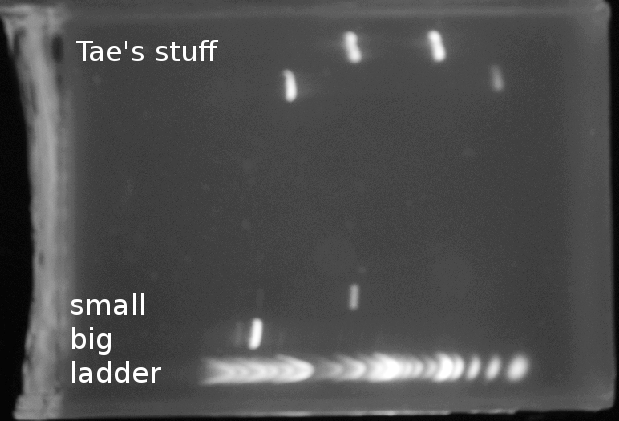
| 6, 7, 8 | Tae's stuff |
- Ran gel at 160 volts
- Started at 12:00 PM
- Stopped at 12:30 PM
- Imaged under long blue light:
- Cut out my 8 bands (four large-fragment bands; four small-fragment bands)
- Placed all large-fragement bands into new tube labeled
KL big plasmid - Placed all small-fragment bands into new tube labeled
KL small plasmid.
- Placed all large-fragement bands into new tube labeled
- Added 600μL ADB buffer to each tube.
- Heated tubes to 55°C and vortexed to melt bands.
- Placed tubes in freezer.
Note: I expected one 4kb fragment and possibly one 40bp fragment from the digest gel purification. However, I could not clearly determine the sizes of the bands due to insufficient separation in the gel. Therefore I saved both sizes of fragments.
2014-04-03
- Retrieved vector plasmid digest fragments
KL big plasmidandKL small plasmidfrom freezer. - Finished Zymo cleanup on each tube; eluted with 30μL ddH2O.
- Created 10-well gel consisting of:
- 5g agarose powder
- 50mL distilled H2O (this was accidental)
- 5mL 10x TAE buffer
- 5μL GelGreen dye
- Loaded wells as follows:
| Lane | Contents |
| 1 | 5μL ladder |
| 2 | 2μL KL big plasmid + 6μL ddH2O + 1μL loading dye
|
| 3 | 2μL KL small plasmid + 6μL ddH2O + 1μL loading dye
|
- Ran gel at 160 volts
- Started 10:58
- Stopped at 11:55
- Imaged gel under trans UV light:
- Stored big and small plasmid digest fragments in freezer with labels
KL big plasmidandKL small plasmid.
Note: I still could not clearly read the sizes of the large and small digest fragments. Upon comparison with my teammates' digest gels, I determined that the large fragment should be correct. The small fragment tube KL small plasmid will be discarded.
It also seems a bit odd that the large fragment lane in the gel shows several fainter bands.
2014-04-08
- Retrieved plasmid digest
KL big plasmidand insert digestsT pure, 4KLandV pure, 4KLfrom freezer. - Setup ligation two ligation reactions, one for each insert:
| 6.5μL | ddH2O |
| 1μL | T4 DNA ligase buffer |
| 1μL | plasmid digest KL big plasmid
|
| 1μL | insert digest T pure, 4KL or V pure, 4KL
|
| 0.5μL | T4 DNA ligase |
- Mixed contents, let sit on benchtop.
- Started 10:14
- Ended 10:44
- Began transformation:
- Obtained one 200μL cell aliquot; kept on ice.
- Added 30μL KCM to cell aliquot.
- To each finished ligation tube, added 70μL cell mixture; discarded cell mixture.
- Let the two tubes sit on ice for 10 minutes.
- Heat shocked between 46°C and 50°C (used IncuBlock, which does not have precise temperature control)
- Started 10:55
- Ended 10:58
- Added 100μL LB media, put on IncuBlock to incubate
- Began 11:17
- Ended 12:11
- Temperature varied between 36°C and 41°C.
- Plated cells onto ampicillin plates:
- 100μL of T construct on plate
T 4KL - Over 100μL of V construct on plate
V 4KL
- 100μL of T construct on plate
- Discarded empty ligation reaction tubes
- Put plates into 37°C incubator for overnight incubation
- Started 12:23
Notes: If the colonies don't turn out well, then controls will be run. Negative control: transform the digested vector plasmid alone (should not get any colonies). Positive control: transform original vector plasmid (should get colonies).
2014-04-10
- Retrieved T construct and V construct plates. Got no colonies. Discarded plates.
- Running positive control
- Obtained 200μL cell aliquot
- Added 30μL KCM
- Mixed 70μL cell mixture and 2μL pBAD OLED plasmid (borrowed from Chris Coates) into new tube.
- Heat shocked at 42°C for 3 minutes
- Added 100μL LB media
- Incubated at 37°C starting 11:15, ending 12:10
- Plated
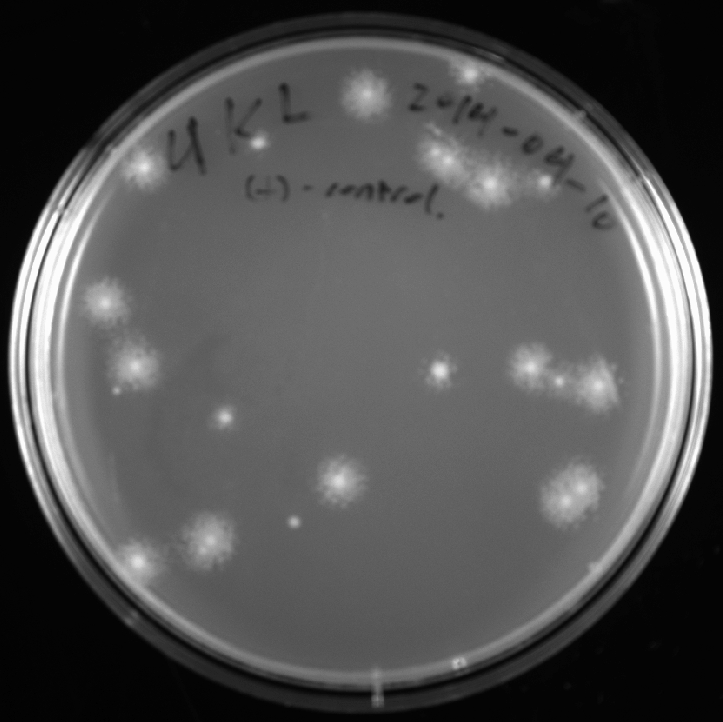
2014-04-11
- Retrieved positive control plate. Imaged under EPI white light:
- Sealed plate with parafilm and stored in refrigerator.
Note: positive control worked. So next step is to retry ligation with equal mass of insert and vector (i.e. equal brightness of gel bands).
2014-04-15
- Retrieved positive control plate.
- Put 4mL LB/ampicillin liquid media in each of two new test tubes.
- Labeled tubes
4KL pick 1and4KL pick 2
- Labeled tubes
- Picked two colonies and put in respective tubes:
- Stored test tubes in warm room.
- Re-sealed plate with parafilm and stored in refrigerator.
- Conducted inventory check:
| label | contents |
TA, TB, VA, VB |
SOE PCR intermediate products for T and V constructs |
T pure, V pure |
T and V construct digests |
T pdt, V pdt |
unknown (!) |
4KL 2014-04-10 (+)-control |
colonies with pBAD OLED plasmid (originally miniprepped by Chris Coates) |
2014-04-17
- Retrieved test tubes
4KL pick 1and4KL pick 2from refrigerator.- Tubes are cloudy
- Miniprepped 2mL of cells from each tube and obtained two 50μL tubes of DNA.
- Labeled plasmid DNA tubes:
4KL OLED pBad colony 1and4KL pBad OLED colony 2
- Labeled plasmid DNA tubes:
- Donated test tubes of cells to Christy Truong
2014-04-22
- Retrieved tube
4KL OLED pBad colony 1from freezer. - Performed two identical 30μL digests (called "digest 1" and "digest 2"). In each digest:
| content | quantity |
| plasmid | 15μL |
| 10x NEB buffer "2" | 3μL |
| ddH2O | 9μL |
| EcoRI | 1.5μL |
| NcoI | 1.5μL |
- Incubated at 37°C
- Started 10:17AM
- Ended 11:10AM
- Added 2μL DNA gel loading dye to each digest product
- Loaded three right-most wells of a 10-well gel:
| lane | contents |
| 8 | 12μL digest 1 |
| 9 | 6μL digest 1 and 6μL digest 2 |
| 10 | 12μL digest 2 |
- Ran gel at 160 volts
- Started 11:22AM
- Ended 12:10PM
- Visualized gel:
- Cut three bands, put into three zymo columns, and performed zymo cleanup
- Eluted each of the three columns with 10μL ddH2O
- Final plasmid digest label:
4KL plas dig
2014-04-24
Attempted to find relative concentrations of plasmid digest and insert digest by running gel with 1μL of each. However, the resulting bands were too faint to visualize.
2014-04-25
Attempted to ligate and transform with 1μL of plasmid digest and 2μL of insert digest, but no cells were available. Ligations were abandoned.
2014-04-29
- Setup two ligations:
| ddH2O | 5.5μL |
4KL plas dig |
1μL |
T pure, 4KL or V pure, 4KL |
2μL |
| T4 DNA ligase buffer | 1μL |
| T4 DNA ligase | 0.5μL |
- Started ligation at 10:08 AM. Ended 10:42 AM.
- Got 200μL cell aliquot; added 30μL KCM and 50μL ddH2O
- Put 70μL cell mixture into each ligation mixture.
- Gave remainder of cell mixture to Tae
- Heat shocked and incubated at 37°C
- Started 10:58 AM
- Ended 12:00 PM
- Plated the two transformations on two halves of a plate.
- used between 100μL and 200μL for plating
2014-05-01
- Retrieved plate from 2014-04-29 from refrigerator.
- Imaged under white light:
- Got a mess of colonies on V construct side only. No colonies for T construct.
- Moving ahead with V construct only. Abandoning T construct.
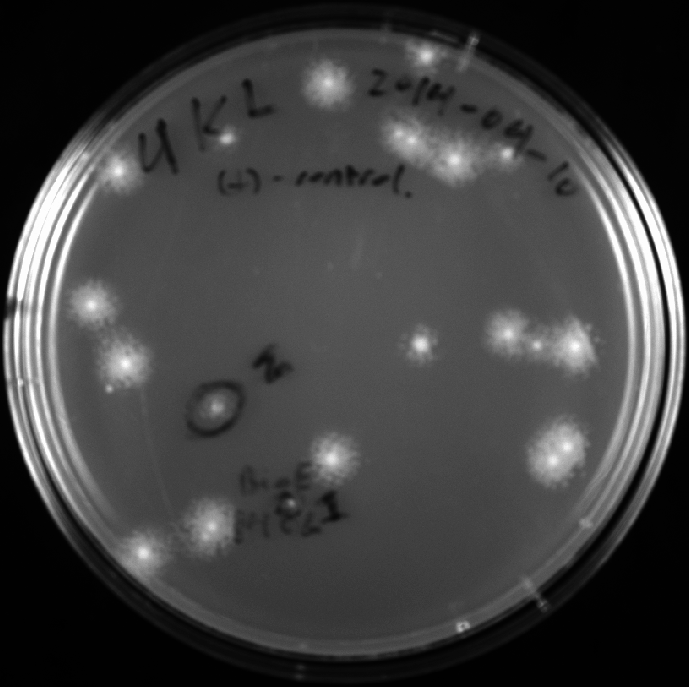
- Picked two colonies from V construct.
- Put in new tubes
4KL V pick 1and4KL V pick 2with 4mL LB/ampicillin media - Imaged colony pick locations:
- Put in new tubes
- Put tubes in 37°C incubator. Wrapped plate in parafilm and stored in refrigerator.
2014-05-02
- Retrieved tubes
4KL V pick 1and4KL V pick 2from incubator.4KL V pick 1is cloudy4KL V pick 2is clear
- Proceeded to miniprep with only
4KL V pick 1.- Did 2 identical minipreps, totalling less than 4mL of cells.
- Eluted plasmid DNA with total of 100μL ddH2O.
- Eluted into tube
4KL V miniprep
- Conducted restriction mapping
- Digested both
4KL V miniprepand pBad plasmid4KL OLED pBad colony 1 - Used BamHI
- Digested for 90-120 minutes.
- Digested both
| content | volume |
| DNA | 15μL |
| NEB buffer 2 | 1μL |
| BamHI | 0.5μL |
- Created 10-well gel with:
- 0.3g agarose powder
- 30mL 1x TAE
- No GelGreen because we ran out (big mistake!!)
- Loaded wells as follows:
| lane number (from left) | contents |
| 1 | blank |
| 2 | blank |
| 3 | BamHI digest of 4KL OLED pBad colony 1 (10μL) + loading dye (1μL)
|
| 4 | 2-log ladder (5μL) |
| 5 | BamHI digest of 4KL V miniprep (10μL) + loading dye (1μL)
|
- Ran gel 160 volts
- Started 4:34 PM.
- Ended approximately 5:00 PM.
- Bands are really faint due to lack of GelGreen.
Conclusions
The parent plasmid used throughout the semester was pNE2001 OleD. It is 5307 bp circular and has one BamHI restriction site.
The valyl-tRNA synthetase construct plasmid is, in theory, 6653 bp circular and has two BamHI restriction sites. The theoretical product upon digestion by BamHI would have two fragments: 222 bp linear and 6431 bp.
Thus, the BamHI restriction mapping should show one 5.3 kb band for OleD plasmid and two bands (6.4 kb and 222 bp) for valyl-tRNA synthetase plasmid. But I didn't get that. Instead, I got one band for valyl-tRNA synthetase plasmid and that fragment is shorter than the OleD band. Something is really wrong.
Note that it is possible I got more bands, but since the gel wasn't made correctly, only the brightest bands are visible in the image. Note that even the ladder is really faint.
The threonyl-tRNA synthetase construct didn't get any colonies after ligation and transformation. Due to lack of time, this construct was abandoned in the last week.