~~!~~
Friday
| The Plan
|
The Day
|
- Re-seed in morning for large scale comp cells.
- make large scale comp cells!
- Re-do all the transformations! Remember controls!!!
|
|
~~!~~
Thursday
| The Plan
|
The Day
|
- Pick in the morning. Remember controls!!
- Decided to forgo making large scale comp cells from things that I haven't sequenced. I will make comp cells from my transformation today of my sequenced Bsrs051 into MC1061.
- Check Tecan results
- Prepare PP for MM.
- Re-seed in proper conditions for ON (tecan??)
|
- Tecan worked!
- The LexA rbs variants didn't work. They comp cells I did the library into were clearly just contaminated with broken reporter...
- Picking from Bsrs051 in MC1061 (also picking from comp cells I used yesterday just in case) Picking into 2YT CA
|
Wednesday
| The Plan
|
The Day
|
- Check Sequencing
- Pick (remember controls!)
- Re-seed for ON and Tecan
- Today, I'm going to make comp cells from Bsrs051 plate.
- I'm also going to transform Bsrs051 (clone 1) into MC1061 to make a new plate.
- I'm also going to pick to make ON culture to make comp cells tomorrow.
- Today I will transform in my Bsrs049 rbs clones into the comp cells as well as Bsrs050.
- I will continue with the Tecan, but not rely on the data.
- Transform Bsrs049's and Bsrs050 into Bsrs051 comp cells
- Transform Bsrs051 into MC1061
|
- Everything sequenced perfectly except rbs7 which looked like it had a huge insertion at the rbs. So don't use it.
- Plates are f*cked. I need to remake the comp cells... from mini-prep.
- Transformed Bsrs049 (rbs 3, 5, & 8) (forgot Bsrs050!) into both the Bsrs051 comp cells
- Transformed Bsrs051 into MC1061
- Recovered
- Plated on CAS, plated on CA (also plated plain comp cells on CA and plated comp cells on Spec for contamination check)
|
Tuesday
| The Plan
|
The Day
|
- Pick
- Mini-Prep, sequence, Transform, plate
|
- Mini-prepped all the new LexA408 rbs members (3-8)
- I transformed all the clone 1's into both Bsrs051 and MC1061, plated on CAS and Spec respectively
- I sent in all clones for sequencing
- I picked two colonies of each constructu: 1) MC1061 2)Bsrs051 (normal) 3) Bsrs051 (broken?) 4-7) Bsrs050, 49.1,49.2,38 in MC1061 8-11) Bsrs050, 49.1,49.2,38 in Bsrs051 comp cells
- These are located in two 24 well blocks because i screwed up the MC1061 constructus (MC1061 are in their own)
- NOTE: Bsrs051 comp cells might be contaminated... but more likely it looks like the promoter 'broke' because it has the CA resistance but is white, but does not grow on Spec alone. That is why I picked both 'normal' (green) and 'broken?' (white) to see growth affects.
|
Wednesday
| The Plan
|
The Day
|
- Take end-point measurement on my ON from the libraries
- Mini-Prep best, and plasmid separate
- Check Tecan results
|
- Tecan results looked really good; I got a 11-12 fold reduction in GFP with -/+ AHL with 1ul into 200ul.
- Redo these Tecans with sula(408/408) alone in CA, Constructs alone in Spec, Reprter with Dummy plasmid again, and white cells for comparison
- End point measurements showed clones 1, 5, 6, 14, 18, & 24as the lowest normalized RFU.
- I mini-prepped these and they will be of mixed plasmids.
- will do a plasmid separation transformation of all 6 into MC1061 and plate on CA
|
Tuesday
| The Plan
|
The Day
|
- Check sequencing
- Make Large Scale Comp cells. Come in at 7:30 to Put 5ml of overnight into 500mL of 2YT.
- Pick Library members and grow in CS
- Tecan overnight stuff
|
- Bsrs051-1 was perfect. Bsrs051-2 had a single point mutation that changed a Lysine -> Threonine in the sfGFP. It doesn't seem to have had an effect in the RFU...
- I have to use Bsrs051-2 in the Tecan, but I'll pick from only the Bsrs051.1's (next week) to redo the expeirment. I will also only use Bsrs051-1 for comp cells.
- The library plates looked really good. Nice diversity and a lot of completely off white cells which shows that the double mutant reporter an the LexA408 is working. I picked 24 colonies; 6 from each combination of rbslibrary and reporter.
File:20120124 13158(crop).png
you can see very nice healthy completely white cells as well as healthy green.
- Made large scale comp cells for Bsrs051, and marked them with purple and put them in the drawer second from the bottom
- Also, did an overnight Tecan with LB, DMSO, & DMSO+AHL
|
Monday
| The Plan
|
The Day
|
- Mini-Prep reporters and sequence
- Make more comp cells, and digest, ligate, transform in LexA408 library into new reporters. Plate on CAS
- Pick from yesterday's plates, and grow in correct media conditions.
|
- Started re-seed for comp cells and ON to make large scall comp cells tomorrow
- Digested Pcon-C5 and LexA408 rbs library with Bam/xhoi and BgIII/xhoi respectively.
- There are no BsaI sites in my Pcon-lexA408 construct so I can move my system over the analogous system that team cI is using.
- Sent in the mutant mutant promoters for sequencing
- Made double mutant comp cells
- Digest, Ligated, Transformed rbslib.LexA408 into double mutant comp cells
- plate on CAS
- Picked 2 colonies of each transformation (except 2.38 & 2.49.2 because they were redundant and not useful) and grew in LB CS
|
Easter Sunday
| The Plan
|
The Day
|
- Re-seeding to mini-prep and sequence confirm tomorrow
- Making comp cells from both Bsrs051 reporters
- Transforming in Full-LexA408 & LexA-TraR
- Recover
- Plate on CAS
|
- Re-seeded the Bsrs051 ON's to make new ON and to make Comp cells
- Mini-prepped (will sequence tomorrow/ in Scott Box Under Construction)
- Made comp cells from Bsrs051 clone 1 and 2
- Doing four transformations into each. Bsrs038, Bsrs49.1, Bsrs049.2, and Bsrs050. Should be green, white, white, and green respectively.
- All will be plated on CAS
|
Saturday
| The Plan
|
The Day
|
- Pick
|
- Picked two green colonies into CA
|
Friday
| The Plan
|
The Day
|
- Analyze Sequencing
- Eco/Bam DIgest PCR product of new reporter
- Eco/Bam 1601CA maybe some other 1601... 1601c? 1601a?
- Gel Purify
- Ligate
- Transform
- Plate
|
- multiple nhe1 sites explain weird ass bands from yesterday. Luckily one of the LexA-TraR constructs ligated correctly So it can be used.
- (Reminding myself to ligate the rbslib.LexA with Ptet and no-nhe1 Pcon's)
|
Thursday
| The Plan
|
The Day
|
- Mini-Prep
- Map, Sequence
|
- There was only one red culture of 3 for both Trunc5 and Truc17. The background (Ptet-cI-rbsRFp) should be red, and so should mine since its on a Pcon, so there should be no white colonies. Strange... checking band sizes. My LexA construcuts should be 200 bps less than the cI constructs. (TraR being ~1000 & 1200; RFP being around 1150 & 1350)
File:20120406 14110(small).png
- Clearly some mixed ligations, but I'm sequencing 1, 2, 5, 6, 8, 10 & 11.
|
Wednesday
| The Plan
|
The Day
|
- Pick
|
- Picked 3 colonies each into LB Spec
|
Tuesday
| The Plan
|
The Day
|
- Check digest band sizes
- ((((BgIII/xhoI Digest library members of rbs.LexA408 # Bam/xhoI Digest 9525-Pcon; wait to have 408/408 reporters))))
- Nhe1/Eco 9525-Bsrs049 PCR Truncations
- Nhe1/Eco TraR, LambdaRep, MukF, ER? (pick large/plasmid band)
- Gel Purify
- Ligate
- Transform
- Recover, Plate
|
- Mini-prepped some 9525 cI fusions for Nina; then digested the TraR fusion and RFP drop out fusion
- Digested:
File:20120404 14080(small).png
- It looks faint because the Eth. Bromide was old, but should be high concentration.
- Started Ligation of T5 & T17 with both TraT and rbs.RFP
- Transformed into MC1061 and plated on Spec
|
Wednesday
| The Plan
|
- MM notes:
- create a 408/408 reporter
- Test a homo system with LexA408-TraR (this will be easy and should take about a week) & see if you can get AHL dependancy
- In meantime see if you can just KO LexA (cell viability) (LexA KO's are not viable. they will not grow.)
- Also, make sure that my system isn't demonstrating OD growth defects or toxicity; Ask gabe/postdocs how to gauge for toxicity
- Get off Spec Plasmids (do in parallel in 9145 or something, not 9525); change reporter to just Cam... maybe? Anything except Spec, and not Amp if we're doing 9145... (Use K, C, & A)
|
Spencer is on Spring break 3/22-4/2
Wednesday
| The Plan
|
The Day
|
- Mini-Prep the Bsrs049's and QC
- Scrape library members; Mini-Prep
- Ask Robin & Austin to Mini-Prep more 9525-1834 / Two vials please!
- DESIGN OLIGOS (over spring break; study for your exam first)
|
- Mini-Prepped the Bsrs049's and the scraped libraries.
- Sequenced well (throw away 1.1 and 2.1) 2.21 is the same as 2.17 but lacks the silent mutation so might be better to use. really shouldnt matter though.
|
Tuesday
| The Plan
|
The Day
|
- Check QC
- Pick two colonies (white!); grow in spec.
- PCR for truncation
- PCR cleanup
- Digest
- Ligate
- Transform; Plate
- Design oligos to give normal LexA the same rbs
- Design Oligos for KO's and such.
|
- Everything was good except the clone 1 which was really weird.
- Picked two colonies of each and grew in Spec
- Did some PCR's off the 5 and 17 clone for truncations and a re-do of the rbs library
- the Truncations failed because I used to wrong digestion; its okay because I can make these in parallel with the w.t. LexA later
- The rbs library's are gel purified and are ligating into 9525
- Will Transform into MC1061, recover, and Plate with beads on Spec
|
Monday
| The Plan
|
The Day
|
- Re-seed clones 1, 5, 17, 21, and controls for ON
- Re seed same clones for Tecan
- Mini-Prep said clones
- Send for Sequencing with special primers
- Design Oligos for KO's and such.
|
|
Sunday
| The Plan
|
The Day
|
- Re-seed for more overnights
- Re-seed into Tecan
- 24 Mini-preps
- Send for QC! with our special oligos
- Plasmid separate somehow?
- Maybe transform all of them even though we don't know QC stuff
|
- Re-seeded for more overnights
- Re-seeded to set up a Tecan
- Mini-prepped only clone 1 & 5 because they looked promising
- Will send for sequencing on Monday
|
Saturday
| The Plan
|
The Day
|
- Pick 24 white cells
- Run colony PCR?
- Make/Use new CAS media
|
- Picked 24 colonies and grew in CS
|
Friday
| The Plan
|
The Day
|
- Scrape
- Mini-prep
- Digest
- Ligate with Pcon's
- Transform into Bsrs048 comp cells! (Remember Controls!, dummy 9525 plasmid on CAS and regular comp cells on CA)
- Plate on CAS
|
- Scraped but only used 1/4 of the pellet to make the mini-preps
- Digested Pcon (clone 2 & 3) with Bam /xhoi (1480+1000, S)
- Digested Bsrs038 with rbs lib with BgIII/xhoi (2100 + 900, L) (could definately see background bands from 1834, but I think I got the right bands. Background will be at 3300 & 900)
|
Thursday
| The Plan
|
The Day
|
- Digested PCR's
- Purified, & Ligated into 9525
- Transformed and Plated on Spec
|
- Digested PCR's
- Purified, & Ligated into 9525
- Transformed and Plated on Spec
|
Wednesday
| The Plan
|
The Day
|
Re-seed for more overnights
- Re-seed into Tecan
- 24 Mini-preps
- Send for QC! with our special oligos
- Plasmid seperate somehow?
Maybe transform all of them even though we don't know QC stuff
|
- Need to re-think plan. I have to KO both LexA and sulA/sulA promoter!
- In the meantime I might just carry on with cloning to make the mutants and truncations...
- Nevermind! I can get around this and use the genomic LexA to my advantage in the Bsrs038 strains.
- Started SOEing on 9525-rbslib-LexA constructs to make the 408 mutant so that i can transform them into Bsrs038 cells.
- Did two rounds of SOEing. I ran it on a gel and the bands look good but could be brighter so I'm doing what Nina suggested and doing one more round of PCR just to amplify the final product. I will pick that up tomorrow.
|
Tuesday
| The Plan
|
The Day
|
- Pick white colonies! (if the controls check out)
- Make more CAS plates
|
- Not as white as I would hope for but compared to normal white cells some are alright.
- ganna pick 24 and do Col PCR on them all
- There were 126 colonies in the 1:1000 dilution and 90ul of plating.
- Ran Col PCR Ran on Gel: All worked except 2 that had mixed bands!
 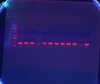
|
Monday
| The Plan
|
The Day
|
- Bam/Xhoi Digest 9525-C5
- BgIII/Xhoi Digest 9525-(rbs.lib)-bsrs038
- Purify
- Ligate Two separate batches
- Transform into MC1061 & SulA reporter strains
- Include controls
- Plate on Spec & CAS
|
- Started large scale comp cell prep of Bsrs047 and Bsrs048 in 1601CA
- Digested Pcon (clone 2 & 3) with Bam /xhoi (1480+1000, S)
- Digested Bsrs038 with rbs lib with BgIII/xhoi (2100 + 900, L) (could definately see background bands from 1834, but I think I got the right bands.)
- Ligated
- Transformed into my new Bsrs037 comp cells. With a 9525-Pcon dummy control. Also plated the comp cells just on Spec to check for contamination.
- Recovering
- Plated 30ul of 1x, 90ul of 1/100, & 90ul of 1/1000
- Need to make more CAS plates
|
Sunday
| The Plan
|
The Day
|
- Pick
|
- Everything sequenced well (except one Pcon) Good to go!
|
Saturday
| The Plan
|
The Day
|
- Mini-Prep
- Map; Sequence
- Sequence Pcons
- Transform; Plate
|
- One of each SulA reporter clone didn't grow. The ones that did grow weren't very high OD. I'm going to pick more if the mapping doesn't look good.
- Mini-prep'd
- E/B digested bsrs047/048 will map
- Transformed bsrs047/048 will plate on CA
- Sent all the Pcons and bsrs047 & bsrs048 in for sequencing
- Reporters mapped well.
|
Friday
| The Plan
|
The Day
|
- Pick
- Mini-Prep
|
- Scrape both plates of the 9525-(rbs.lib)-Bsrs038 only in the concentrated parts
- Ask Gabe how to do it again…
- Mini-Prep
|
Thursday
| The Plan
|
The Day
|
- Change everything
- Transfer reporters to 1601CA
- Start construction of Pcon-rbs-LexA
|
- E/B Digested Bsrs047 & Bsrs048 and 1601CA.
- Gel Purify smaller bands
- Ligate Bsrs047 and Bsrs048 into 1601CA
- Ligate Pcon-C5 into your pre-digested 9525
- Transform into MC1061
- Plate on CA and on Spec respectively
|
Wednesday
| The Plan
|
The Day
|
- Make Bsrs047 and Bsrs048 Comp Cells
- EIPCR rbs's onto my two LexA constructs
- Map, E/B Digest & E/B digest 9525 dropout or backbone, Map
- Purify
- Ligate
- Transform into MC1061; also transform into SulA comp cells
|
- Sequencing was perfect for all reporter constructs
- Started Bsrs047 and Bsrs048 comp cell cultures in 2YT-Spec at 8:48am
- Did 6 total Expand EIPCR's with the '55' thermo program on 9145-Bsrs038, 9525-Bsrs038, & 9525-Bsrs044 (2 on each) (should be done around 11:15am)
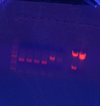
- Digested; Picked lanes 1, 3, 5, 7 & 8
- Purified, Ligated 1 with 8 and 5 with 7
- Transformed, Recovered, Plated on Spec full plates
|
Tuesday
| The Plan
|
The Day
|
- Send in mini-preps for sequencing
- transform in mini-preps to create comp cells
|
- Sent for sequencing; Trasnformed all 4 into MC1061 and plated on Spec
|
Spencer Is in New Zealand 2/24-3/6
Friday
| The Plan
|
The Day
|
- Pick/ Grow
- Ask someone to Mini-prep on Saturday
|
- Gabe mini-prepped for me
|
Thursday
| The Plan
|
The Day
|
- Check Sequencing
- If oligos come in, ask how long PCR reaction stay good; and PCR LexA constructs to add rbs's
- Also see how long colonies on a plate last, and potentially digest, ligate, transform, plate
|
- Great sequencing! Got all of our parts
- Bam/xhoi& BgIII/xhoi digested 1600-Bsrs045/Bsrs046& 1600-jtk2541:
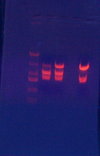
- (1486, 1227, S) & (2239, 1149, L)
- Ligated; Transformed into MC1061; Recovered; Plated on Spec
|
Wednesday
| The Plan
|
The Day
|
- mini-prep, map, sequence
- BgIII/Brick Assembly of SulA reporters with JTK2541
- PCR LexA constructs to add rbs's!
|
- The shaker wasn't at 37... so nothing grew up. Picking into 2YT and will mini-prep in ~6-8 hours.
- 24 mini-preps!
- 24 Eco/Bam Digestions!
- Lengths: 1-4: (2635, 753); 5-10: (2066, 615); 11-16: (2481, 615); 17-24: (2481, 1108)
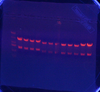 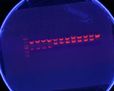
- They all worked except the last one. Sent in for sequencing 1, 2, 8, 9, 10, 11, 12, 14, 17, 19, 21, & 22.
|
Tuesday
| The Plan
|
The Day
|
- Ligate JTK2541 into 1600 & 1601CA; Ligate LexAFull(w.t) into 9145 & 9525
- Ligate both LexA(1-87)(w.t.)'s into 9525-nhe1.rbs.RFP
- Transform into MC1061, Recover, Plate on Spec, CA, Amp, Spec, Spec respectively
- Come in late at night to pick Colonies! Grow in respective media. ~12 hours later
|
- Five Ligations:
(1) JTK2541&1600
(2) LexAFull & 9145
(3) LexA Full & 9525
(4) LexA(1-87).1 in 9525
(5) LexA(1-87).2 in 9525
- Transformed everything into MC1061, recovered, plated.
|
Monday
| The Plan
|
The Day
|
- Map PCR cleanup's of LexA (before and after digestion)
- Re-do Ligations of PCR cleanup's of LexA into 9145 & 9525; maybe try a p15? And 1600
- Do we need to do chasis modifications?
- Eco/Bam JTK2541 into 1600 and 1601CA
|
- The PCR's of LexA look good. I will try to re-digest and re-ligate them.
- Need to move JTK2541 into 1600 and 1601CA so I can ligate it with SulA promoters.
- Eco/Bam, JTK2541, 1600, 1601CA, 9145, 9525, & LexA (w.t)
(1) (2481+753, S)
(2) (2635+78, L)
(3) (3205+78, L)
(4) (2066+342, L)
(5) (2481+1854, L)
(6) (615)
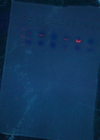
- Eco/Nhe1 LexA(1-87)'s and 9525-1834
(1) 261
(2) 261
(3) (3313+1022, L)
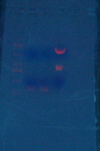
- Gel Purify
|
Sunday
| The Plan
|
The Day
|
- Check Sequence Results
- Do some EIPCR on LexA w.t. (and maybe the trunc if it sequenced well!)
- PCR cleanup
|
- The SulA Promoters assembled in both 1601CA and 1600. However, none of the LexA constructs worked. They were all really strange.
- Sequence the LexA PCRs??
- Need to somehow get LexA!
|
Saturday
| The Plan
|
The Day
|
- Check Sequence Results
- mini-prep SulA promoter cells;sequence
|
- Sequences sucked. The assembly went wrong, somehow only the stress-ToxR was left in most of them. Bss003 and Bss006 sequences well at least. Sent the other potential Bsrs044 constructs in for sequencing along with 14 other sulA promoter sequences and some LexA w.t sequences.
|
Friday
| The Plan
|
The Day
|
- Re-seed into Arabinose titration Tecan and run overnight.
- QC 9145-Bsrs041 cells with pBgl00001-Bsrs043 & pBgl00001-Bss006 (mini-prep them)
- Mini-prep 9525-LexA constructs; Map; Sequence
- Pick SulA promoter cells; grow in CA; Pick 9145-Bsrs038 cells (not red!) grow in AMP
- Re-Sequence Bss003 & Bss006
|
- Picked 4 colonies for each SulA promoter grew in CA for the 1601CA and Spec for the 1600.
Also picked four non-red 9145-Bsrs038 cells, grew in AMP.
- Set up the titrations
- Eco/Bam'd the 8 Lex(1-87) to map. (desired size: 2400,1100)
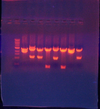
Lanes 2, 3, 5, & 8 looked promising; I realized that they shouldn't be red because there is not promoter, so anything that was red like lane 1 is most likely background; i sent for sequencing since I want to have this part anyway.
- Sent for sequencing 1, 2, 3, 5, 8, Bss003, Bss006, and the mixed plasmids reading from ca56
|
Thursday
| The Plan
|
The Day
|
- Pick from 9145-Bsrs041 cells with pBgl00001-Bsrs043 (Grow in AT) and with 9525-Bsrs043 (Grow in AS)
- Check Sequencing
- Pick from 9525-LexA constructs (Grow in Spec)
- Wobble PCR SulA promoter (op+/op+) & (408/op+)
- Short Frag Clean-Ups
- Eco/Bam Digests on PCR's and 1601CA
- Gel Purify
- Two Ligations
- Two Transformations, Recover, Plate on CA
|
- Wobbled 189f/190r & 191f/192r (~100bps)
- Small Frag clean-up done.
- Eco/Bam'd the two Wobble reactions and 1601CA-Bss52.
I accidentally put 7 ul of Part for 1601CA instead of 3 so I added .5ul of NEB2 in addition to the original .5, such that it should be around 10% of the volume.
- Sequencing data came back; I forgot to specify primers so the only read I got was ca56 off Bss006 which looked great.
- Gel purified (small frag purified for the sula promoters)

- The 1601CA was kinda weak so I also used a E/Bam digest of 1600 from Gabe that might also be sketchy. I ligated each of the sulA promoters into these plasmids. (4 ligations)
- Transformed into MC1061; Recovered for 30 mins in 2YT
- Plated 1601CA on CA and 1600 on Spec.
|









