Ryan N. Willhite Week 4
From OpenWetWare
Jump to navigationJump to search
In-Class Activity
Activity 1, Part 2
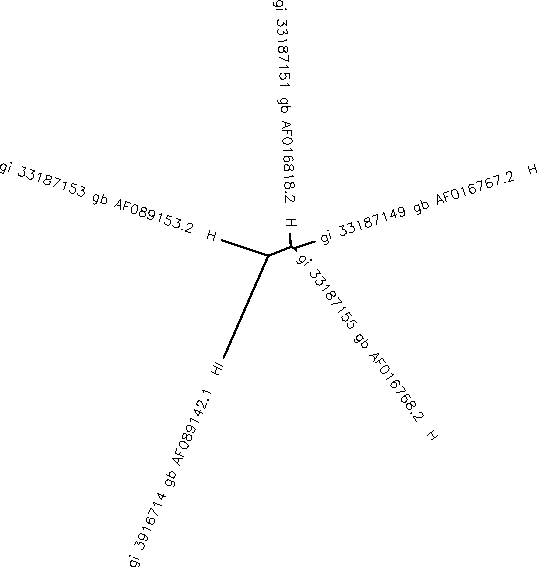
- The accession number from the sequence I chose was AF089153.
- The subject of the study was HIV-1 isolate S4V2-3 from USA envelope glycoprotein. The section of the record that contained information about who the HIV was collected from was in the subject.
- In order to get to the FASTA format, on he NCBI website the displau drop down menu allowed me to view the selected sequences in FASTA format.
- uploaded file as (.txt), it is the FASTA file sequences from downloading sequences to the local hard drive.
- Ran into some trouble opening the FASTA file since there was no type it recognized. In order to overcome this issue, i saved as a .txt file extension and then opened it in notepad.
Activity 1, Part 3
Activity 2, Part 1
- To find first visit info. go to sequence data under Bedrock.
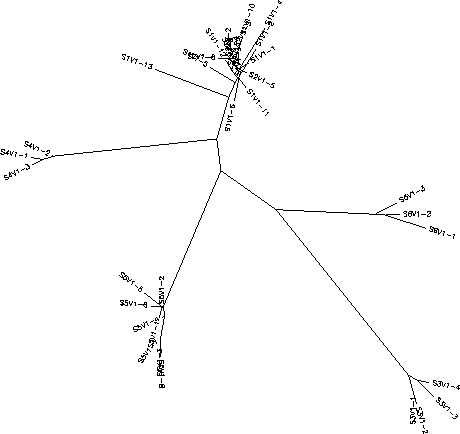
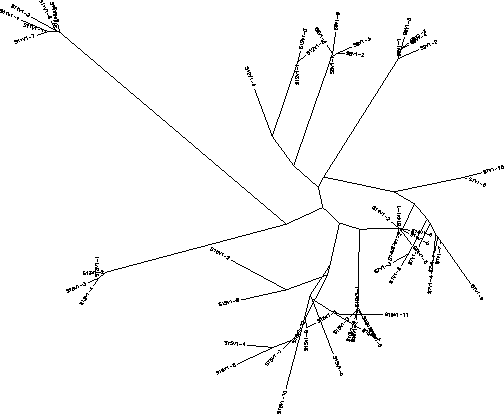
- It gets tight where there is a lot of confusion on the tree because it shows that these particular sequences are much more similar than the others across from them or further in distance.
- Do the clones from each subject cluster together? Yes
- Do some subjects' clones show more diversity than others? Yes
- Do some of the subjects cluster together? Yes
Activity 2, Part 2
This aspect of the activity focuses on ways to quantify sequence similarity and difference. An S statistic is used to quantify the diversity of sequence in a population. This part was extremely confusing and need more help in this area. Mainly, doing a pairwise distance matrix.