Registry of Standard Biological Models/CellML Practical/Practical 1
Goal: Describe a simple genetic assembly (promoter + RBS + protein coding region + stop codon) from the description of each sub-component.
Example of such a part: <bbpart>BBa_I7101</bbpart>
Simple Modeling
Assumptions: > Central Dogma: Gene --> mRNA --> Protein > unlimited resources to produce mRNA and proteins (i.e. excess of Polymerases, Ribosomes, tRNA ...) > No stochastic effect taken into account, continous approach
<amsmath>\frac{d[mRNA]}{dt} = k_M*[Gene] - \gamma_M*[mRNA]</amsmath>
<amsmath>\frac{d[Protein]}{dt} = k_P*[mRNA] - \gamma_P*[Protein]</amsmath>
Where,
<amsmath>\emph [Gene]</amsmath>= number of gene copies per cell, [Gene]
<amsmath>\emph [mRNA]</amsmath>= mRNA concentration per cell, [mRNA]
<amsmath>\emph [Protein]</amsmath>= Protein concentration per cell, [Protein]
<amsmath>\emph k_{M}</amsmath>= mRNA production rate (PoPS), unit:[mRNA]/[DNA]/s
<amsmath>\emph k_{P}</amsmath>= Protein production rate (RiPS), unit:[Protein]/[mRNA]/s
<amsmath>\gamma_M</amsmath>= mRNA degradation rate, 1/s
<amsmath>\gamma_P</amsmath>= Protein degradation rate., 1/s
BioBrick matching
| Component | Function | Related parameters | Comments |
|---|---|---|---|
| Promoter | tanscription | <amsmath>\emph [Gene], \emph k_{M} </amsmath> | the promoter can be characterized with a given rate of transcription rate (PoPs) which depends on its intrinsic nature and the number of gene copies available in the system |
| RBS | translation | <amsmath>\emph k_{P}</amsmath> | the RBS defines the rate of translation per mRNA |
| mRNA | degradation | <amsmath>\gamma_M</amsmath> | the mRNA component is not a BioBrick, however it has a specific degradation rate |
| Protein | degradation | <amsmath>\gamma_P</amsmath> | without considering its function, the protein should be characterized by its half-life. |
CellML matching
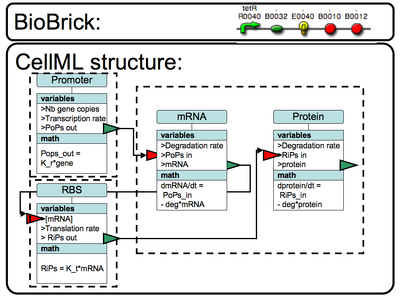 |
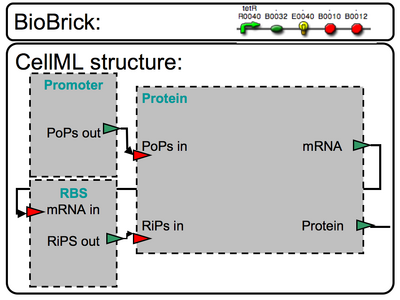 |
>>>>Comments
- About the Promoter:
- Characterized by nb gene copies + transcription rate
- PoPs is defined as a MathML expression. Can easily define repression, activation or constitutive expression behaviour.
- Inputs: none. BC: Should numbers of copies be here? VR: true.
- Output: PoPs (Polymerases per second). It is used to feed a Protein component. BC: Although it feeds into a protein component here, more generally it could feed into other components, such as tRNA. VR: Not too sure to understand how you define PoPs for tRNAs. Can you explain ? BC:I mean that PoPS should be independent of whatever piece of DNA you are sitting on when you measure it. So the polymerases could be transcribing an mRNA or a tRNA or whatever. Since we can easily imagine wanting to transcribe non-protein components such as tRNA, we need to keep the output of the promoter general in my opinion. VR: I am a bit confused. To me polymerases are only binding DNA (chromosomal or plasmid). tRNA are compounds formed by an anti-codon and amino-acid site, to me they onlyinteract with the ribosome during translation of the protein. Am I missing something ?
- About the RBS:
- Characterized by translation rate
- RiPS is defined by a MathML expression. Could describe limited resource at this point.
- Inputs: needs [mRNA]
- Ouputs: RiPs (Ribosomes per second). It is used to feed a Protein component.
- About the Protein:
- I felt the need to encapsulate in the same component the concept of mRNA and protein. mRNA is not a BioBrick anyway. This component might benefit being implemented as a group, or maybe simply an import of a 'protein_molecule' component and a 'mRNA_molecule' component.
- We should think about riboswitch parts which are affecting mRNA behaviour
- Characterized by degradation rate of mRNA and Protein
- Inputs: need PoPs from a promoter + RiPs from a RBS
- BC:Do we really need PoPS as an input to a protein component. I feel that the PoPS defines the numbers of mRNA and after that, it doesn't really affect the protein component. What do you think? VR: I have to admit that I haven't found yet a design with which I am happy. My problem comes from the fact that the mRNA component is not a BioBrick, so I would like to hide it some how. I have also tried to encapsulate it with the RBS component here.
- Outputs: [mRNA] + [Protein]