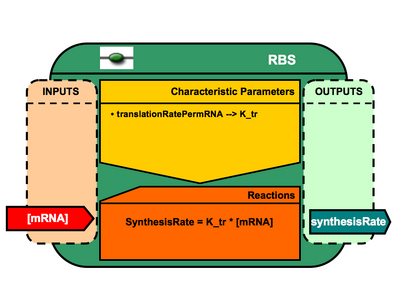
Registry of Standard Biological Models/Basic Component Models/Ribosome binding site (RBS)
ID: Ribosome Binding Site (RBS)
CellML structure (CellML 1.1 spec)
- Component: RBS
- Units:
- Imported from Environment component
- Variables:
- mRNA (public interface= in )
- translationRatePermRNA (public interface = none / init value = XXX)
- protein-synthesis-rate (public interface = out / init value = XXX)
- MathML
- <amsmath> proteinSynthesisRate = translationRatePermRNA*[mRNA]</amsmath>
CellML File
<syntax type='xml'> <?xml version="1.0"?>
<model xmlns="http://www.cellml.org/cellml/1.0#"
xmlns:cmeta="http://www.cellml.org/metadata/1.0#" xml:base="file:///C:/CellML_models/RBS.cml" cmeta:id="RBS" name="RBS">
<component name="RBS">
<variable name="proteinSynthesisRate" initial_value="" public_interface="out" units="moles_per_second"/> <variable name="nb_mRNA" initial_value="" public_interface="in" units="mole"/> <variable name="proteinSynthesisRatePermRNA" initial_value="1" public_interface="none" units="per_second"/>
[math]\displaystyle{ \lt apply id="proteinSynthesisRate"\gt \lt eq/\gt \lt ci\gt proteinSynthesisRate\lt /ci\gt \lt apply\gt \lt times/\gt \lt ci\gt proteinSynthesisRatePermRNA\lt /ci\gt \lt ci\gt nb_mRNA\lt /ci\gt \lt /apply\gt \lt /apply\gt }[/math]
</component>
<import xmlns:xlink="http://www.w3.org/1999/xlink"
xlink:href="file://C:\CellML_models\units.cml">
<units name="moles_per_second" units_ref="moles_per_second"/>
<units name="per_second" units_ref="per_second"/>
</import>
</model>
</syntax>
Comments
- this component provides the synthesis rate of downstream proteins based on [mRNA] and RBS translation rate.
- don't know if it is possible to characterize experimentally the translationRatePermRNA?