Registry of Standard Biological Models/Basic Component Models/Promoter RBS Coupled
Promoter RBS Coupled Architecture
|
 |
CellML structure (CellML 1.1 spec)
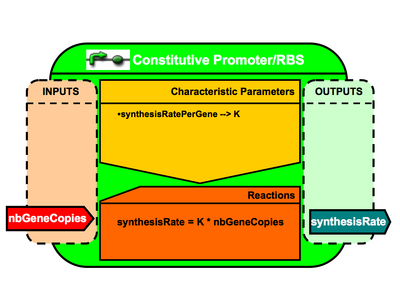
- Component: PromoterRBSCoupled
- Units:
- Imported from Environment component
- Variables:
- nbGeneCopies (public interface = in / init value = 1.0)
- RiPSOutPerGene (public interface = none / init value = XXX)
- RiPSOut (public interface = out / init value = depends on the promoter-RBS used)
- MathML
- [math]\displaystyle{ RiPSOut = nbGeneCopies*RiPSOutPerGene }[/math]
Examples
| BioBricks from Registry | CellML file |
|---|---|
| <bbpart>BBa_J13211</bbpart> ==> TetR (<bbpart>BBa_R0040</bbpart>) + RBS (<bbpart>BBa_R0032</bbpart>) | Media:BBa_J13211_Model.xml |
| other part | other file |
CellML File
<syntax type = 'xml'>
<?xml version="1.0"?>
<model xmlns="http://www.cellml.org/cellml/1.0#"
xmlns:cmeta="http://www.cellml.org/metadata/1.0#" xml:base="http://openwetware.org/index.php?title=Registry_of_Standard_Biological_Models/Basic_Component_Models/Promoter_RBS_Coupled/CellML_Code&action=raw" cmeta:id="promoter-RBS_constitutive" name="promoter-RBS_constitutive">
<component name="promoter-RBS_constitutive">
<variable name="RiPSOut" initial_value="" public_interface="out" units="moles_per_second"/> <variable name="nbGeneCopies" initial_value="1" public_interface="none" units="dimensionless"/> <variable name="RiPSOutPerGene" initial_value="1" public_interface="none" units="moles_per_second"/>
[math]\displaystyle{ \lt apply id="RiPSOut"\gt \lt eq/\gt \lt ci\gt RiPSOut\lt /ci\gt \lt apply\gt \lt times/\gt \lt ci\gt RiPSOutPerGene\lt /ci\gt \lt ci\gt nbGeneCopies\lt /ci\gt \lt /apply\gt \lt /apply\gt }[/math]
</component>
<import xmlns:xlink="http://www.w3.org/1999/xlink"
xlink:href="http://openwetware.org/index.php?title=Registry_of_Standard_Biological_Models/Basic_Component_Models/Units&action=raw">
<units name="moles_per_second" units_ref="moles_per_second"/>
<units name="per_second" units_ref="per_second"/>
</import>
</model>
</syntax>
Comments
- This component is simply a container providing the synthesis rate specific to a constitutive promoter coupled to a RBS
- Having a model of a promoter and RBS coupled could mean that we have a constant synthesis rate of the downstream protein(s) (quasi steady state approx)
- This parameter is easier to characterize experimentally. Using a calibrated GFP assay with none half-life, we know that at steady state (synthesis = degradation)