Olivas Lab:Research
Our Research
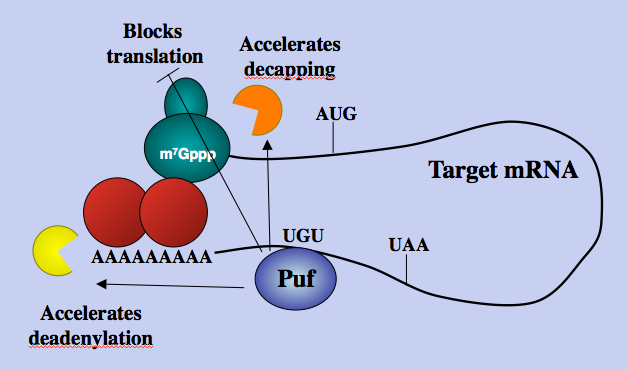
Cells respond to environmental signals and stresses by transcriptional regulation of response genes, as well as activation of more rapid post-transcriptional regulatory pathways. Modulation of mRNA degradation and translation rates is an efficient method to rapidly alter protein production in response to cellular changes. The Puf family of eukaryotic RNA-binding proteins control critical decisions in cell development and differentiation by regulating the degradation and translation of target mRNAs through interactions with 3’ untranslated regions (UTRs). Pufs are also involved in cellular stress responses. Research in our lab utilizes the yeast model system Saccharomyces cerevisiae to determine how Puf proteins integrate environmental signals to alter their regulation of mRNA metabolism. For example, we have demonstrated that Puf3p responds to environmental carbon source signals to regulate the decay of nuclear-encoded transcripts that code for mitochondrial proteins. Specifically, Puf3p stimulates mRNA decay of such transcripts in fermentative growth conditions (in glucose) when mitochondrial respiration is not required. In contrast, Puf3p activity is inhibited in ethanol, galactose and raffinose conditions that utilize mitochondria, thereby stabilizing the mitochondrial transcripts for upregulated translation. The rapid changes in Puf3p activity in response to carbon source are due to post-translational mechanisms that do not alter the ability of Puf3p to bind mRNA. In addition to our work with Puf3p, we also have data showing that the yeast Puf proteins 1, 4 and 5, which play a role in nitric oxide stress response and ribosome biogenesis, are also differentially regulated in response to environmental conditions. Our current work focuses on understanding the mechanisms controlling the activity of these Puf proteins.
To complement our mechanistic studies in yeast, a second project in our lab is studying mRNAs in human neural cells that are targeted by Puf protein regulation. It is hypothesized that Puf proteins and miRNAs act cooperatively to regulate certain human mRNAs, so our studies are also investigating this activity.