Microfluidic Bacterial Analysis - Joshua Gukowsky
Introduction
Bacteria are a topic of great interest to scientists due to their central role in biological systems and human health, but their miniscule size can make them a challenge to study. Using a microfluidic device to aid in bacterial research can help to improve the accuracy and efficiency of the analysis, and can facilitate the exploration of more novel areas of study such as individual cell analysis[1] and the study of chemotaxis (cell movement).[2] Microfluidic-assisted bacterial analysis is a growing area of research with the potential to help significantly enhance our collective understanding of these organisms and how they impact our health. Improving the speed and accuracy of bacterial analysis can also help to identify the causes of bacterial disease, and potentially save lives by helping to prevent their spread.
Methods of Bacterial Analysis

There are a number of microfluidics-based techniques that can be used to analyze bacteria, which can be separated into several categories for different types of samples. Microfluidic bacterial analysis can be performed with either single-phase systems, in which bacterial suspensions are directly flowed through channels, or droplet-based systems in which small volumes of the bacterial culture are manipulated in the device while suspended in an immiscible liquid.[4]
Bacterial Culture Analysis
It is possible to analyze cultures of bacteria using microfluidic devices to assess the characteristics of large numbers of cells in a single sample. Examples of applications of microfluidic devices in this area include the study of bacterial cell growth in cultures under different conditions,[5] determining the antibiotic susceptibility of bacterial cultures,[6] and the extraction of DNA from a sample of cells.[7] In these applications, the bacterial culture can either be grown separately and then analyzed in a device, or cultured directly in a microfluidic system containing appropriate growth media. There are distinct considerations to take into account when growing a bacterial culture within a microfluidic device, depending on the type of organism and growth media being used. One such consideration is the availability of oxygen, and whether or not the organism is aerobic or anaerobic. PDMS, a material commonly used to make microfluidic devices, has a high gas permeability and cells inside a PDMS device will be able to grow aerobically.[8] In order to grow cells anaerobically within such a device, it would be necessary to store the device in anaerobic conditions or potentially coat or modify the PDMS to reduce the permeability of oxygen. Another potential concern is the surface characteristics of the device- PDMS, for instance, is hydrophobic and has been shown to negatively impact the growth of certain types of cell cultures.[9] The surface of the device can be modified to have more favorable growth conditions through decreasing its hydrophobicity or coating it in specific substances, such as fibronectin, to assist with cellular adhesion or growth.
Single Cell Analysis
The characteristically small scale of microfluidic devices makes it feasible to study individual bacterial cells. To facilitate the study of single cells, it is typically necessary to include a mechanism in the device capable of trapping or isolating individual bacteria. Existing studies have demonstrated several effective mechanisms to trap single bacterial cells, with varying degrees of effectiveness. An example of trap that can be used is the “sieve” design developed by Kim et al.,[10] shown in Figure 1. This design contains a cup-like structure with a ~1 μm gap to help guide and capture individual bacteria. While this trap design was effective for analyzing single cells, it exhibited a fairly low cell capture rate- only about 1% of the cells in the starting culture were successfully trapped. This suggests that this kind of trap design would be less effective for analyzing a sample containing a low number of cells, since a higher bacteria concentration may be necessary to get a sufficient number of cells trapped for subsequent analysis.

An alternative method for capturing bacteria is to use a porous filter membrane, which can be incorporated into a microfluidic device.[12] The bacterial culture can be flowed through the membrane, trapping the bacteria on the filter. The cells on the filter membrane can then be analyzed by removing the filter from the device, or the device may be designed to allow the filter to be accessible from the exterior, allowing it to be directly analyzed without taking the device apart.
Biofilm Analysis
A different method for bacterial analysis that utilizes microfluidic devices is the study of biofilms, which are communities of cells that form complex aggregates and become encased in an extracellular matrix.[13] Biofilms can be formed directly on surfaces within a microfluidic device, which eliminates the need for traps or filters to capture the bacterial cells and can make them easier to study than individual organisms. Biofilms can pose a considerable threat to human health, both by forming in the human body (such as plaque that forms on teeth) or by forming in locations where they could lead to human infections, such as on medical devices or food processing equipment. Microfluidic devices can be used to study bacterial biofilms in a number of different ways, and can facilitate the research process by reproducing the biological processes that lead to their formation on a small and economical scale. An example of one such application is the use of microfluidics to study the formation of biofilms in simulated dynamic environments, replicating conditions such as seawater in which nutritional bioavailability and the surrounding chemical conditions are highly heterogeneous and variable.[14] Microfluidic devices can also be used to study specific properties of biofilms, such as their susceptibility to different antibiotics[15] and their ability to respond to flow in controlled conditions [16].
Applications
Microfluidic bacterial analysis is a growing area of study that is helping to expand our understanding of these organisms and improve our ability to detect and characterize them. Four of the main applications that these technologies have been used for are pathogen detection, the determination of antibiotic susceptibility, the study of bacterial physiology, and biotechnological alteration or improvement of bacterial strains.[17]
Pathogen Detection
Microfluidic devices can be used to facilitate the detection of pathogens in tandem with chemical detection methods, such as fluorescence labeling or spectroscopy-based techniques. For instance, the surface of the device can be modified with specific antibodies to capture a target bacterial organism that can be subsequently identified using fluorescence.[18] Optical detection of pathogens on a microfluidic device is also possible- Park et al.[19] utilized a smartphone app to quantify the degree of agglutination of antibody-conjugated particles when exposed to a pathogen in a paper microfluidic device.
Antibiotic Susceptibility
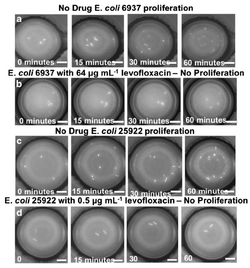
A simple application of microfluidics to characterize bacteria is the determination of antibiotic susceptibility, which can be accomplished relatively fast on a small scale. Li et al.[21] designed a method for determining minimum inhibitory concentrations for multiple antibiotics within 8-14 hours, which is an improvement on the typical timescale (2 to 4 days) required to obtain these results. Other microfluidics-based methods for determining antibiotic sensitivity include using fluorescent biosensors to track the proliferation of small numbers of cells[22] and using spectroscopic techniques to assess antibiotic susceptibility of captured bacterial cells.[23] Identifying cellular resistance or susceptibility to specific antibiotics in a rapid manner is necessary for efficient treatment of bacterial infections, and can reduce the prescription of unnecessary antibiotics[24] and ultimately help prevent the further development of antibiotic resistance.
Bacterial Physiology
Beyond antibiotic susceptibility, microfluidic platforms can be used to assess other physiological characteristics of bacteria. Examples of potential applications include studying bacterial chemotaxis,[25] analyzing the structural integrity of bacterial cells,[26] and determining the biochemical characteristics of viable-but-non-culturable (VBNC) organisms.[27]
Biotechnology
Another application of microfluidics is the small-scale manipulation of biochemical processes to assist with biotechnology applications. For instance, DNA can be amplified in microfluidic devices in a simplified and efficient manner,[28] which can then be utilized for further applications such as gene recombination and transformation. Microfluidic systems can also be used to study enzyme expression and for protein analysis.[29]
Challenges and Future Research
Microfluidic microbial analysis is an increasingly large and diverse area of study, and researchers in this area face several challenges that will need to be overcome in the future. These challenges include simplifying complex preparation and handling procedures, and improving the accessibility of these technologies to a wider audience.[30] Future microfluidic systems for bacterial analysis may make use of automated technology and allow for improved precision of experimental parameters (such as reagent concentration). Creating accessible technology that can be easily and inexpensively utilized in commercial applications will also help to increase the usage of microfluidic devices and improve upon the existing methods used for commercial microbial analysis.
References
- ↑ https://pubs.rsc.org/en/content/articlelanding/2011/lc/c0lc00362j#!divAbstract
- ↑ https://www.sciencedirect.com/science/article/pii/S1872204017610508
- ↑ https://pubs.rsc.org/en/content/articlelanding/2011/lc/c0lc00362j#!divAbstract
- ↑ https://pubs.rsc.org/en/content/articlelanding/2016/lc/c6lc00367b#!divAbstract
- ↑ https://pubs.rsc.org/en/Content/ArticleLanding/2014/LC/C3LC51332G#!divAbstract
- ↑ https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3489182/
- ↑ https://www.ncbi.nlm.nih.gov/pubmed/20938545
- ↑ https://link.springer.com/article/10.1023/A:1024583026925
- ↑ https://www.sciencedirect.com/science/article/pii/S0956566314005302
- ↑ https://pubs.rsc.org/en/content/articlelanding/2011/lc/c0lc00362j#!divAbstract
- ↑ https://www.ncbi.nlm.nih.gov/pubmed/25959709
- ↑ https://www.ncbi.nlm.nih.gov/pubmed/17388566
- ↑ https://pubs.rsc.org/en/content/articlelanding/2012/lc/c2lc20800h#!divAbstract
- ↑ https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5019069/
- ↑ https://pubs.rsc.org/en/Content/ArticleLanding/2010/LC/c0lc00154f#!divAbstract
- ↑ https://academic.oup.com/femsle/article/312/1/33/470352
- ↑ https://pubs.rsc.org/en/content/articlelanding/2016/lc/c6lc00367b#!divAbstract
- ↑ https://pubs.rsc.org/en/content/articlelanding/2015/lc/c5lc00971e#!divAbstract
- ↑ https://www.ncbi.nlm.nih.gov/pubmed/24162816
- ↑ https://link.springer.com/article/10.1007%2Fs00604-017-2492-9
- ↑ https://www.ncbi.nlm.nih.gov/pubmed/30609582
- ↑ https://link.springer.com/article/10.1007%2Fs00604-017-2492-9
- ↑ https://www.ncbi.nlm.nih.gov/pubmed/28464521
- ↑ https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6190172/
- ↑ https://www.sciencedirect.com/science/article/pii/S1872204017610508
- ↑ https://www.ncbi.nlm.nih.gov/pubmed/29266097
- ↑ https://bmcbiol.biomedcentral.com/articles/10.1186/s12915-017-0465-4
- ↑ https://pubs.rsc.org/en/Content/ArticleLanding/2013/LC/C3lc41097h#!divAbstract
- ↑ https://pubs.rsc.org/en/content/articlelanding/2016/lc/c5lc01039j#!divAbstract
- ↑ https://pubs.rsc.org/en/content/articlelanding/2016/lc/c6lc00367b#!divAbstract
