Measuring Bacterial Gene Expression
Introduction
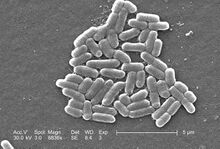
Escherichia coli is a rod-shaped bacterial species commonly used in cell culture studies. E. coli is favorably used as a model organism because its genome is well understood and it can be easily grown in laboratories (at ideal conditions, E. coli divides every 20 minutes) [1]. Additionally, gene expression in E. coli can be easily controlled to select for certain mutant strains (e.g. antibiotic resistance) [1]. Thus, E. coli has gained use as a host cell for production of recombinant proteins after its genomes is altered via a plasmid vector [2].
Gene Expression
Measuring gene expression develops a better understanding of metabolism and cellular signaling pathways. E. coli is an ideal organism for studying metabolism due to its inherent simplicity (making it easier to locate sites of interest). In cell culture, expression is measured by the type and quantity of proteins/ions that are produced following treatment.
Technical Limitations
Classic cell culture studies the growth and activity of a variety of cell types, ranging from mammalian cell lines to bacterial colonies. However cell culture is constrained by plate or suspension culture, which has several drawbacks. Some of the major constraints stem from the dependence on petri dish culture, including (but not limited to): the lack of an ability to address and integrate three-dimensional (3D) cell-to-cell interactions, the lack of control over exposure to media and the subsequent exhaustion of nutrients in plate media, the lack of stability in the colonies as time progresses, and the dependence on averaging results due to the prevalence of dense colonies.
Microscale Measurement of Gene Expression
Microfluidic devices are apparatuses predominantly made out of polydimethylsiloxane (PDMS) layers on silicon wafers that allow for exquisite control over miniscule fluid volumes. Through various microfluidic techniques (e.g. photolithography), microfluidics has introduced a means to reliably and effectively create and control variables and environments that can be readily replicable.
Benefits
Microfluidics introduces a level of control and customization that is not limited to the averaging of many colonies in two-dimensional plane. Now, the field can replicate in vivo structures, study cell-to-cell interactions in a three-dimensional space, and observe processes on a single-cell level of specificity [3]. Microfluidic devices designed for cell culture are as simple as only being a single layer of PDMS. Exquisite control of the microenvironment can also establish devices that have gradients to measure different nutrient concentrations over a range, as seen in chemotaxis with varying pH levels in different streams.
Challenges
It is important to prevent cross-contamination of separately-treated cells (e.g. individual cells exposed to different types of media). Additionally, excessive shear stress could induce mechanical stress on cells and interfere with measurement of cellular response (when response should only be dependent on treatment of cells) [4]. There must be an effective means of separating individual cells when they divide; flowrate can prevent cellular adhesion and formation of colonies so that cells can be measured individually. The video to the right demonstrates an effective means of trapping individual cells; with each cellular division, the progeny gets trapped in a successive well within the channel[5].
Applications
Biomedical Research
Microfluidic devices have potential for stem cell research. Since microfluidic devices have microenvironments (e.g. pH) that can be controlled, changing these conditions can influence response of stem cells (e.g. proliferation rate, differentiation) [3]. Establishing concentration gradients, as seen in chemotaxis and durotaxis gradient devices, can measures stem cells response under a variety of conditions.
Pharmaceutical Research
Microfludic devices enable high throughput for drug testing by only needing to administer them in minute quantities. Isolation of cells in these devices can be done with ease, as well as using channels to direct drugs to cells and measure their response to the drug [3].
References
- ↑ 1.0 1.1 Rosano, G. L. & Ceccarelli, E. A. Recombinant protein expression in Escherichia coli: Advances and challenges. Front. Microbiol. 5, (2014). http://dx.doi.org/10.3389/fmicb.2014.00172
- ↑ RMakrides, S. C. Strategies for achieving high-level expression of genes in Escherichia coli. Microbiol. Rev. 60, 512–+ (1996). https://doi.org/10.1128/mr.60.3.512-538.1996
- ↑ 3.0 3.1 3.2 Makrides, S. C. Strategies for achieving high-level expression of genes in Escherichia coli. Microbiol. Rev. 60, 512–+ (1996). https://doi.org/10.1128/mr.60.3.512-538.1996
- ↑ Rusconi, R., Garren, M. & Stocker, R. Microfluidics Ecpanding the Frontiers of Microbial Ecology. Annual Review of Biophysics, Vol 43 (ed. Dill, K. A.) 43, 65–91 (2014). https://www.annualreviews.org/doi/full/10.1146/annurev-biophys-051013-022916#_i36
- ↑ Kimmerling, R. J.; Szeto, G. L.; Li, J. W.; Genshaft, S. W.; Payer, K. R.; de Riba Borrajo, J.; Blainey, P. C.; Irvine, D. J.; Shalek, A. K.; Manalis, S. R. A microfluidic platform enabling single-cell RNA-seq of multigenerational lineages. Nat. Commun. 7, 10220 (2016). http://dx.doi.org/10.1038/ncomms10220
