Macrofluidics for Tumor Metastasis
Macrofluidics: Modelling Tumor Metastasis
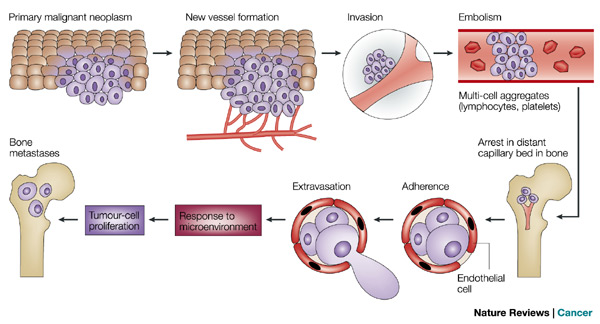
Metastasis is defined as secondary malignant growths that develop at some distance away from the primary site of cancer. This occurs when a primary malignant neoplasm invades a local blood vessel and gets carried by convection to a different location in the body (Figure 1). Once in the bloodstream, these cells are classified as circulating tumor cells. Under certain conditions, these circulating cells can extravasate from the blood vessel into a new environment and begin forming a secondary tumor. More specifically, cancer cells metastasize by degrading local extracellular matrix (ECM) proteins, which allows them to break away from the primary tumor and breach the ECM. However, metastasis does not occur randomly. Certain cancer types preferentially seed into specific organs, as observed by breast cancer favoring lung and bone environments over other organs. The factors governing this trophic activity are not well understood, and current research aims to investigate these factors.
Tumor metastasis accounts for 90% of cancer-related deaths and is characterized by a universally poor prognosis; 5-year overall survival rates drop from 72% for locally advanced cancer to 22% once metastatic [1]. Currently, metastatic cancer has only been successfully modeled in costly in-vivo models and, to some degree, using mathematical systems unsupported by empirical evidence. Novel research is being conducted with the goal of developing a quality in-vitro device capable of modelling tumor metastasis in a simulated microenvironment, with focus specifically on the phenomena of tropism, cancer's predisposition to colonize particular tissue. An estimated 1.7 million new cases of cancer are expected to be diagnosed in 2016, with all cases of cancer doubling worldwide by 2032 [1].
Current Methods for Modelling Cancer Metastasis
In-vivo Animal Models
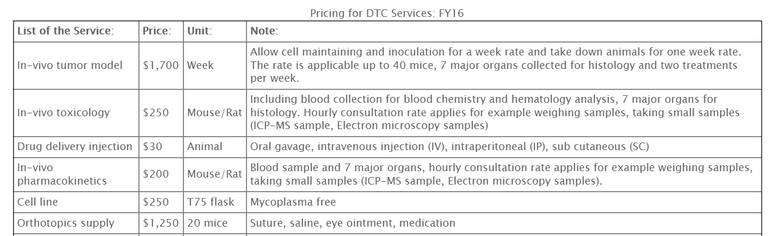
Animal models are the current standard for all measures of cancer testing, with particular emphasis on mouse models. As it stands, mice are the most cost efficient in terms of space and handling, and they provide a biological system comparable to humans. AntiCancer Incorporated [?] has developed an efficient way for laboratories to obtain highly specialized mouse models with their customizable service, MetaMouse [2]. MetaMouse offers researchers the ability to order mouse models with a predetermined cancer type grafted or cultured in a tissue or organ of interest. The service boasts of "more than 100 GFP/RFP expressing tumor models" in which "all types of agents, small molecules, proteins, genes, can be evaluated" [2]. Because animal models are complex, it can be difficult to develop experimental procedures that isolate variables of interest. Performing tests in mice can also be cost prohibitive, as the average base cost for maintaining an in-vivo model is $1700 per week (Table 1). Although studying tumor metastasis in animal models is a viable option, we argue that moving away from this approach to anin vitro model would prove even more effective.
Tumor Transplantation
Several methods exist for generating metastasis models in mice canonical to human cancer: tumor transplantation, genetic modification, and the amplification of specific promoters. Perhaps the most pervasive technique is one where cancer cells of a particular phenotype with specific tropic factors can be grafted into host mice to develop a model. Human cancer cells can be xenograft into mice for the purpose of determining what factors allow a patient's cancer to persist and metastasize. This technique allowed researchers to "implicate a number of genes in organ-specific metastasis, including IL-11, osteopontin and the connective tissue growth factor (CTGF) in osteolytic metastasis," which generated a method to specifically modulate the factors affecting these genes and increase the efficacy of targeted treatments [3].
Genetic Modification
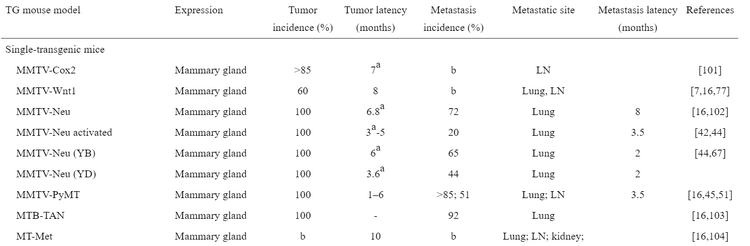
Specific genes have been implicated in the generation of cancers with particular tropic tendencies. This has allowed researchers to modify a genetic sequence or multiple genes to generate a mouse model whose cancer would form in a particular tissue and metastasize to a known region. Through the use of a Mouse Mammary Tumor Virus (MMTV), two genes have been identified as primary targets for generating malignant tumors. Researchers have determined that MMTV-Neu and MMTV-PyMT transgenic mice develop cancer tropic to the lungs and lymph nodes [3]. This method also allows for the development of multiple different models depending on the desired tissue of study, with one additional example being that "C3(1)-SV40 T-antigen transgenic mice develop invasive mammary carcinomas," making the technique malleable to a variety of purposes.
Drosophila Signaling Pathway
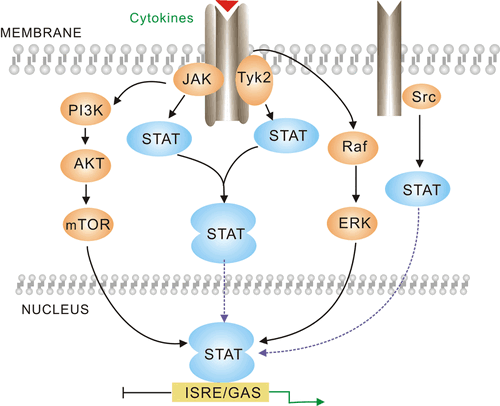
Fruit flies offer a highly tractable animal model which has allowed researchers to develop information on cell signalling pathways, including the JAK/STAT transduction cascade which is responsible for cell growth and has been implicated in tumorogenesis. Drosophila present a canonical pathway for the JAK/STAT cascade which has been conserved by evolution, making the model ideal for performing research related to human patients [4]. This has allowed researchers to develop an understanding of the role the JAK/STAT pathway plays in allowing tumors to form and persist, presenting the opportunity to develop targeted therapies for malignant cancers. Fruit flies are a cost effective in-vivo model and are capable of existing in large quantities, making them easier to maintain than mice colonies.
Mathematical Metastasis Models
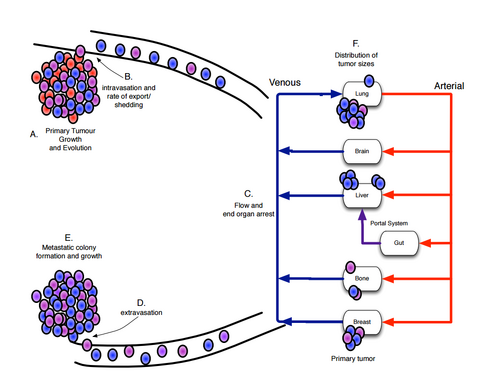
Mathematical models for studying cancer have been employed since 1932, with the initial focus being on understanding tumor growth laws. In current research, math models are used to compliment empirical research by coalescing a wide variety of experimental data into a concise framework of study. Scientists at the Lee Mofit Cancer Center and Research Institute in Tampa Florida cite the power of mathematical models in cancer research as "allowing for identification of the parameters to which the system is the most sensitive, and also allow for logical reasoning beyond what experiments can provide" [4]. These researchers have made efforts to boil down complex biological models into systems of ODEs and PDEs, complete with stochastic properties that mimic the randomness of true biology. Currently, this research has been successful in modelling parameters such as metastatic potential, patterns of spread, and distribution of cancer size. Additional work is still necessary as this is a project in its infancy, however the coupling of these theoretical models with laboratory research have ironed out many of the poorly understood factors effecting cancer metastasis.
Challenges of Current Methods
In-vivo and mathematical models are powerful tools for studying cancer metastasis, but they are not without their drawbacks. In-vivo models suffer from having many inherent variables that are often difficult to control, making it impossible to limit the variables down to the specific areas of interest. In-vitro models offer the ability to knock down all extraneous genes, but to achieve the same effect in animal models would be fatal to the subject. Colonies of animal models are also costly to maintain, with in-vivo toxicology screens costing $250 per rat. This limits the number of studies that can be performed in parallel. Mathematical models are laden with the issue of being complex and difficult to derive, and also do not often have experiments that can be performed to qualify the theoretical systems. Rapid testing using in-vivo models is also a challenge, as living creatures cannot be scaled down beyond their biology.
Macrofluidic Device for Tumor Metastasis
Transitioning to Microscale
There are several benefits to transitioning tumor metastasis studies to a microscale platform. First and foremost, microscale studies will prove to be significantly cheaper than using in vivo models. Although 100mg of PEG maleimide costs upwards of $1000, this cost is greatly offset by the small volume required to construct a single device (http://www.jenkemusa.com). Smaller volumes of materials also means less, waste, so operating at this scale is much more environmentally friendly. With capillary vessels existing on order of 7 μm - 1mm in diameter. working on the microscale makes intuitive sense.
Organ on a Chip
Organ on a chip (OC) is a recent 3D microfluidic device that aims to simulate entire organ systems on the microscale with the purpose of performing bio-microelectromechanical systems (Bio-MEMS) research. Currently, the device has the capacity to human physiology of a heart, lung, kidney, artery, bone, cartilage, and skin. Scaling artificial organs down to the microscale allows for precise cellular manipulation and accurate microenvironment simulation. The advantage that OC has over typical 3D models is the ability to efficiently transport and distribute soluble factors which allows researchers to mimic tissue-to-tissue interfaces, which larger models fail to do. This is due to the transport phenomena associated with the microscale, which allow for effective control of spatiotemporal gradients of chemicals and completely laminar flow which lends itself to cell motility, chemotactic stimuli, and biochemical signalling.
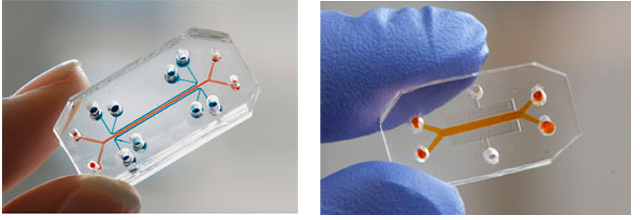
Creating the Macrofluidic Device
A macrofluidic device can be developed in order to model the tumor environment and observe tumor metastasis. The device is made up of a square base structure with a channel connected to two wells in the middle of this structure. A design of the device components and its various dimensions can be developed by using a 3D modeling software such as Inventor. The device base structure is to be created by using a 3D printer to print out the structure using ABS plastic. Original dimensions of the device were made to be 1.5x1.5 inches for the square base and 0.3 inches for the channel and wells. But from tried attempts, it was seen that the 3D printer has limitations in terms of printing out small scale objects and materials. Small scale objects were seen to be printed with less precision and accuracy in terms of the layering of the structure. Since this was the case, the dimensions of the device were scaled up with the square base being 2x2 inches and the dimensions for the channel and wells being three times as big. It was seen and proven that with larger dimensions, the device could be printed and with more precision and accuracy.
Once the macrofluidic device base structure has been created through 3D printing, a PDMS mold will be made off of this base structure. This PDMS mold can be created by first making a 10:1 PDMS mixture solution. Once the mixture solution was made, it was allowed to sit for two hours in a concealed vacuum chamber. After two hours, the mixture was poured over the channel piece from the device base structure in a small petri dish. It is important to note that the channel is really the only necessary part of the device. The square base structure is just used as support for the channel when it was being printed. So afterwards, the dish with the channel piece and mixture solution was placed on top of a heating plate and heated at a temperature of 65 degrees Celsius for about twenty minutes or until the mixture solution has solidified around the channel device. When heating the channel device, make sure to observe that the channel is not floating up in the mixture solution as it would disrupt the formation of the mold. Once the mixture has solidified around the channel device, the PDMS mold is created by simply using a razor blade or knife to carve out the channel piece and leaving behind a mold of the channel. This PDMS mold of the channel is what essentially will be the macrofluidic device that will be used as a fundamental base in modelling the tumor environment.
Modelling the Tumor Environment Using Collagen
In modelling the tumor environment, collagen will be used to fill the channel in the channel device. The collagen will be set into the channel using a very thin 0.18 mm needle. The needle will essentially create this vascular channel where the collagen will set into place and create this sort of mimicked environment for which the cells can be sustained in. The collagen matrix acts to form within the channel by having its pH be raised to about 7.4 in order for the collagen to gel. The collagen should then also be stiffened with the stiffening chemical, Genipin. Once the collagen is set into the channel, epithelial cells are then seeded into the channels and onto the collagen. As to make sure the cells are seeded properly and there is proper capillary formation, the cells will be observed and analyzed under fluorescent microscopy.
Modelling the Tumor Environment Using PEG-Maleimide
If proven success in using collagen to model the tumor environment, then a further step can be taken with using a PEG-maleimide gel to model the tumor environment in place of the collagen. Using PEG-maleimide to model the tumor environment is still an ongoing research that is being done by Professor Shelly Peyton’s lab at the University of Massachusetts Amherst and so creating this model environment properly and successfully is still in working progress. But theoretically, the PEG-maleimide models the environment by chemically filtering out peptide sequences that represent bone marrow tissue (where cancer usually spreads to during metastasis) through capturing integrin binding sites that are in bone marrow tissue. The PEG-maleimide then encloses the epithelial cells in a 3D environment and the cell phenotypes that drive metastasis are separated by this PEG-maleimide system. Since the tumor environment is being modelled and aspects of the bone marrow tissue are being mimicked, characteristics such as stiffness and elasticity of the marrow tissue will also be attempted to be replicated in vitro in this modelled environment by crosslinking the PEG. [5] The procedure in using the PEG-maleimide to model the tumor environment will essentially be similar to the procedure of using collagen to model the environment where the PEG-maleimide will be set into the channel device and then epithelial cells will be seeded onto the PEG-maleimide. One of the main goals of using the PEG-maleimide to model the environment is to see whether or not the cells can survive long enough on the PEG-maleimide. Cell viability will be observed using fluorescent microscopy. If the cells do keep dying, factors such as pH level of the cells, cell density, infection of the cells, and incubation temperature will be looked into as possible reasons for why the cells are not viable.
Quantifying Success
Success for this project is measured by the ability to seed a large percent of endothelial cells in a collagen channel and have them remain viable long enough to perform tests. Cells must persist for aabout 14 weeks, which is the normal effective range of fluorescent tags for endothelial cells. To measure cell seeding effectiveness, a vascularization fraction will be used, which is measured as fec=Length of unseeded Channel/2*Channel Length. This will produce a ratio of how much of the channel was successfully seeded versus unseeded sections. The goal is to obtain a vascularization factor of 75% or greater. Images will be produced using fluorescent microscopy.
References
[1] "Breast Cancer Survival Rates, by Stage." Cancer.org. American Cancer Society, (2016).
[2] "MetaMouse." AntiCancer Incorporated, (2014).
[3] Fantozzi, Anna, and Gerhard Christofori. “Mouse Models of Breast Cancer Metastasis.” Breast Cancer Research 8.4 (2006): 212. PMC.
[4] Scott, Jacob G., Philip Gerlee, and David Basanta. "Mathematical Modeling of the Metastatic Process." Integrated Mathematical Oncology (2015).
[5] Jansen, L, Peyton, S. “A Hydrogel Platform to Understand Breast Cancer Metastasis to Bone Marrow.” AlChE (2015).
