M465:Biolog Ecoplates
Carbon Source Utilization Profiling
You have learned in other courses about the importance of carbon fixation by autotrophic photosynthetic plants. The inability to make carbon-carbon bonds and, therefore, to utilize carbon dioxide as a carbon source, is problematic for heterotrophic species including humans and all other animals. Fortunately there are bacteria that, like plants, are autotrophic and photosynthetic, although many others are heterotrophic, like us. Unlike us, however, bacteria are extremely diverse in the types of carbon sources they can use metabolically. Bacterial communities both compete and co-operate in utilization of available sources of essential, useable carbon. The health and longevity of the community is dependent on a continuous supply of useable carbon for all its members. Your investigation on carbon source profiling will identify carbon sources that can be utilized by your isolates under the conditions we provide them.
Carbon source patterns using BIOLOG™ Ecoplates
Observing patterns of substrate utilization across isolates can provide evidence of functional capabilities of that microbe in its community context. Additionally, understanding how metabolic substrates can be used in the communities can help us understand the stability or flexibility of that ecosystem. Carbon sources are crucial anabolic raw materials for heterotrophic microbial growth. Microbes vary enormously in their ability to make use of carbon in different forms.
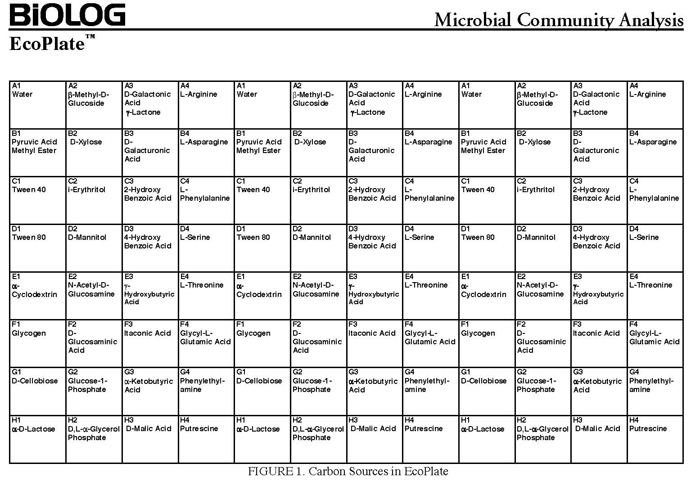
In the following assay, you will use your isolates, inoculated directly into the single carbon source wells of microtiter plates followed by spectrometric quantification of growth. If one or your microbes can use a particular carbon substrate, the metabolism of that carbon source is accompanied by a reduction of water-soluble colorless Tetrazolium salts (WTS) to become reduced purple formazans. WST-1 and in particular WST-8 (2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium (MTT), are reduced outside of the cells. They combine with an electron mediator (phenazine methosulfate (PMS)), to yield a water-soluble purple product called formazan that can be measured spectrophotometrically at 590 nm. The color development is additive and directly proportional to the metabolism of each carbon source so the development of forazan can be followed over time. The intensity of purple color as a pattern in the wells is used to determine the metabolic footprint of your isolate. We will the patterns to determine the metabolic diversity of your isolates (CMD). For these measurements to be meaningful, it is important to control for number of microbes, incubation time, and other microenvironmental factors as well as the requirement for saturating substrate and indicator concentrations. There are 31 carbon sources available on a BIOLOG plate. This set of substrates is far from exhaustive. However, these substrates were chosen for variety and are similar to many nutrients found in natural environments.
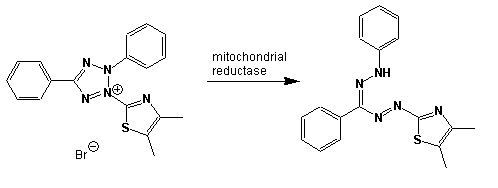
Scheme showing the reduction of MTT to formazan. Image created by Jenpen 21 September 2006
Source http://en.wikipedia.org/wiki/File:Mttscheme.png . Public domain use per Wikipedia Commons.
Colorless (2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium) is reduced to purple formazan
The BIOLOG-ECO™ 96 well plates we will use contain 3 replicates of 31 carbon sources and three water control wells. The method for carbon source profiling that we are using is simple and rapid, but its interpretation must be carefully evaluated, recognizing that the methodology is imperfect. The references below discuss the pitfalls of the ecoplates in the context of community level analyses:
References and Resources: Biolog Carbon Source
• Garland, J.L., Mills, A.L. (1991) Classification and characterization of heterotrophic microbial communities on the basis of patterns of community-level-sole-carbon-source-utilization. Appl Environ Microbiol 57, 2351–2359.
• Garland, J.L. (1997) Analysis and interpretation of community-level physiological profiles in microbialecology. FEMS Microbiol Ecol 24, 289–300.
• Preston-Mafham, J., Boddy, l., Randerson, P.F. (2002) Analysis of microbial community functional diversity using sole carbon source utilization profiles-a critique. FEMS Microbiology Ecology. 42, 1-14.
Lab Activities
Activity #1: Measuring the optical densities of your cultures
Your overnight broth cultures should have some turbidity (growth). We will use optical density at 600 nm as a measure of growth. You will then normalize each of your isolates to the lowest measurement.
1. Vortex your cultures briefly to resuspend the bacterial growth.
2. Label 5, 1.5 mL plastic eppendorf tubes with your isolate number. Label a final, 6th tube, with the word "blank".
3. Using sterile technique, remove 500 uL of broth from each culture into the appropriate 1.5 mL tube. To the "blank" tube add sterile media appropriate to your isolate.
4. To each tube, add 500 uL of distilled, deionized water. You are therefore making a 1:1 dilution. Vortex to mix.
5. Label 6 cuvettes with your name and isolate number or "blank". Take the 1 mL solutions and pipette them into the cuvettes.
6. Using the spectrophotometers provided, measure the optical density at 600 nm (your instructors will show you how).
Activity #2: Carbon source utilization patterns using BIOLOG™ Ecoplates
- Salt Solution (137 mMol NaCl, 2.7 mMol KCl)
- P200 and P1000 micropipets with sterile tips
- Multichannel pipet (set to deliver 100 µL) and sterile tips
- BIOLOG EcoPlate™
- sterile plastic multichannel reservoir
- Wear gloves throughout the entire protocol
- Do not cross contaminate your samples or the solutions
- Keep your work area clean: freshly disinfect your bench top before beginning
- Do not use a vortex at any point in this protocol unless it is specified that you should do so.
Perform all of the below in your laminar flow hood
1. Dilute your isolates, using salt solution, such that they are at an OD600 of 0.1. You will need a total of 10 mL of diluted isolate in salt solution to load your biolog plate. Therefore, if isolate #1 grew to an OD600 of 1.4, you would use C1V1 = C2V2 to calculate the volume necessary to add to a 10 mL salt solution. In this case, I would be calculating: (1.4)*(X) = (0.1) (10mL) and solving for X = 0.714 mL (or 714 uL).
2. You will be using a multichannel micropipet to inoculate your plates with your dilutions. Pour 10 mL of your diluted culture into a sterile reservoir.
3. Be careful to preserve the BIOLOG Eco™ plate's and its cover's sterility (eg. don't place it face down on your bench). Remove the cover and transfer 100 µL of the dilution into each each well of the 96 well BIOLOG plate. Check visually the consistency of the amount of diluted extract in the micropipet tips after you have drawn up your aliquots to determine that you have no bubbles and that the quantity to be dispensed is the same. If the pipet tips appear unevenly filled or you have bubbles, do not dispense the inoculum into the wells! Start over. If you are unfamiliar with the use of multichannel pipets, ask your instructor to observe your technique.
4. Replace the cover of the plate and label one side of the cover. DO NOT LABEL ON the top or bottom to avoid interference in the light passage during spectrophotometric readings. Use a piece of your colored tape and include your initials, the date, and the isolate number.
5. Take a time 0 reading at A590 nm using the BioTek Synergy H1 384 plate reader (your instructors will show you how)
6. After each reading, place your covered back in the incubator (at 30C)
MEASURING MICROBIAL CARBON UTILIZATION AS A590 nm
The intensity of color change is monitored in each of the wells by taking spectrophotometer readings once a day at A590 nm. You will come in daily to collect these data until a peak absorbance is reached on more than 2 consecutive readings, this will likely require daily readings for a week. You must not miss more than 1 consecutive day. Make sure that you take a photo of your plate against a white background on the final day of measurement.
Carbon source utilization pattern:
How do we analyze our data? You could plot on a bar graph the average A590 nm absorbance of the three replicates (with error bars) on the final day of data collection on the y axis and the 31 different carbon sources on the x axis in one figure. (Remember that there is no such thing as a negative value for Absorbance so count anything that is less than zero as zero. Why might you seem to have a negative value?). Should you arrange those carbon sources in the order they are on the BIOLOG plate or is there a better way to organize them on your graph to show your main message(s)? What kind of information do you need in the figure legend?
Can you think of other ways to illustrate metabolic diversity of your isolates?
