Luckau Protocols:STRUCTURE
From OpenWetWare
Jump to navigationJump to search
| STRUCTURE |
Purpose
The software program STRUCTURE is "a model-based clustering method for using multilocus genotype data to infer population structure and assign individuals to populations."
Protocol
- Structure 2.3.3 for MacOS
Formatting Data
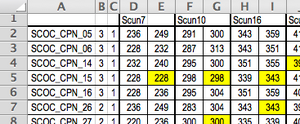
MS Excel
- ROW 1 = marker names (structure term = "Marker Name")
- since diploid, leave an extra column for the second allele between markers
- COLUMN 1 = sample names (structure term = "Label")
- COLUMN 2 = population designation (structure term = "PopData", in integer form)
- Tara's thesis = county area
- 1 = Camp Pendleton
- 2 = Rancho Jamul / Hollenbeck
- 3 = Point Loma / Cabrillo National Monument
- 4 = Santa Ysabel Open Space Preserve
- 5 = Torrey Pines State Natural Reserve
- COLUMN 3 = location designation (structure term = "LocData", in integer form)
- Tara's thesis = array cluster
MS Word
- copy data from Excel, Paste Special into Word
- as Unformatted Text
- Save As ...
- Plain Text (.txt)
- File Conversion: Latin-US (DOS), CR/LF
Structure
New Project
- File: New Project
- Step 1
- Name the project: 20111106AXRJ
- Select directory: Research/Structure/StructureDirectory
- Choose data file: browse to the .txt file you just made (from Excel and Word)
- Step 2
- Number of individuals: 93
- Ploidy of data: 2
- Number of loci: 10
- Missing data value: 0
- Step 3
- Row of marker names - check
- Data file stores data for individuals in a single line - check
- Step 4
- Individual ID for each individual - check
- Putative population origin for each individual - check
- Sampling location information - check
New Parameter Set
- Parameter Set: New...
- Run Length: 10,000 and 10,000
- Ancestry Model
- Admixture
- Independent (we expect allele frequencies in different populations to be reasonably different from each other; works well for many data sets (strong structure))
- Allele Frequency Model: Correlated (frequencies in the different populations are likely to be similar, due to migration or shared ancestry; improves clustering for closely related populations (subtle structure), but may increase the risk of overestimating K)
- Advanced: unclick "Compute probability of data (for estimating K)" to make program run faster
Run Project
- Parameter Set: Run
- K=1 through K=number of sampling sites
- to determine the most likely K,
Literature and Supporting Information
- Pritchard J, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959.
- Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587.
- Falush D, Stephens M, Pritchard JK (2007) Inference of population structure using multilocus genotype data: dominant markers and null alleles. Molecular Ecology Notes 7:574–578.
- Hubisz MJ, Falush D, Stephens M, Pritchard JK (2009) Inferring weak population structure with the assistance of sample group information. Molecular Ecology Resources 9:1322–1332.