Jessica Karen Wong/Notebook/2007-8-8
From OpenWetWare
Jump to navigationJump to search
- Minipreped 2 samples of I2055-SX-B0030 for seq
- Sent both samples (1 was fluor and 1 wasn't) for seq w/ VF & VR
- PCR cleaned I2056 digests (E/X, S/X, both N)
- Ligated I2056-ENX closed, I2056-SNX closed, I2056-EX-B34, I20550EX-B34
- Ligated in the presence of Xba and Spe restriction enzymes: I2055-SX-B32, I2056-SX-B32, I2056-SX-B34, I2055-SX-B34
- Transformed w/ top10
- Ran out of B0032 cut E/S so made an overnight of B0032 to later cut and ligate
- Digested E0240 S/N to go into F2620, prep I2055 S/X, prep I2055 E/X
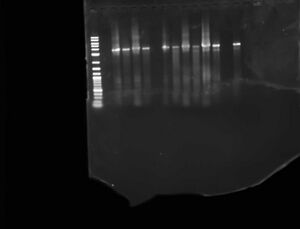
- Ran a gel of the PCR product of each construct next to the Digest of that PCR to make sure digests were cutting in the right place
- Loaded: lad sp I2057-EX, EX Digest, I2057-ES, ES digest, I2056-EX, EX digest, I2056-SX, SX digest, I2055-EX, I2055 EX digest, sp, I2055 SX digest
- Didn't have any more PCR product of I2055-SX
Seq
- I2055-EX seems to have a 1bp mutation at 892 - bad
- I2055-SX has a few bp differences, breaks down towards 806 even w/ VR - bad
- I2057-EX-J116 is actually J102, I2057 part is fine after bp 13
- I2057 first 12 bp's have strange double peaks in chromatogram, large peaks are wrong, small are right
- I2056-7, I2057-ENS, I2057-ENX all failed again
Made overnights of I2055-EX-B0032, I2056 colony 7, I2057-ENS, I2057-ENX, I2055-ENX-2, I2055-SNX-6, I2055-SNX-7 to sequence