IGEM:MIT/2009/pycA Synthesis Plan
Synthesis Plan
Plan for synthesis of the pycA gene and expression in yeast. Please email back if anyone notices any major issue before we send out the order for the synthesis.
Construct
The plan is to synthesis the entire pcyA gene, flanked by the biobrick prefix and suffix so that we can insert the gene into our expression vector as well as directly deposit the part into the registry
[Biobrick Prefix] + [Kozak/RE] + [MTS] + [pcyA] + [RE] + [Biobrick Suffix]

(Genbank .GB File)
Biobrick Prefix - gaattcgcggccgcttctag
Biobrick Suffix - tactagtagcggccgctgcag
MTS - atgcaacgctccatttttgcgaggttc - (Met - Gln - Arg - Ser - Ile - Phe - Ala - Arg - Phe)
pcyA - See below
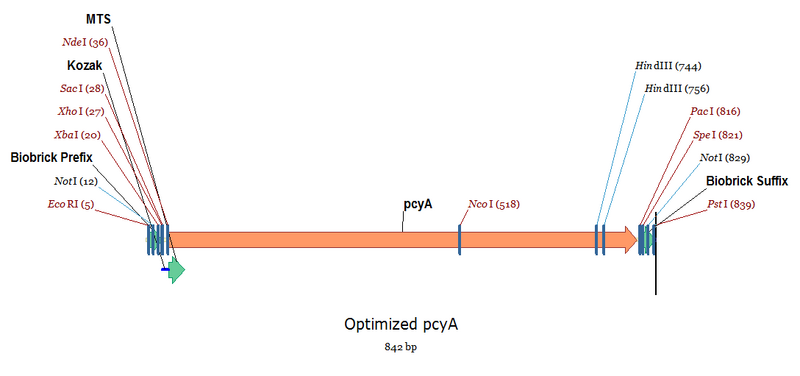
Complete Sequence - Image on the right
This will be ligated into the plasmid YCp22FL1 via XhoI + PacI double digestion (Buffer 4 + BSA). This will not only create our desired sticky ends, but also remove the biobrick bookends. See the annotated sequence on the right for a clearer picture.
Vector
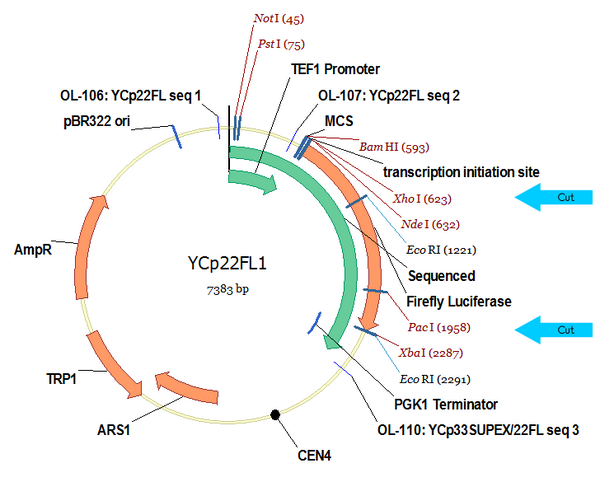
YCp22FL1 (Genbank .GB file)
YCp22FL1 is cut at the Kozak sequence and in the middle of the Firefly luciferase via XhoI and PacI. pcyA is then ligated into this region. We transform this into yeast and then pray.
Non-MTS Construct
Because we also want a version of pcyA without the MTS region, the plan is to design primers that allow for this.
Forward (5'-3')
buf kozak met pcya gcg ctcgagaacat atg gctgttactgatttgtctttgactaattct
Stats
- Melting: 55.7 C
- Worst Hairpin: -1.47 kcal/mol
- Worst Self-Dimer: -9.96 kcal/mol
Reverse (5'-3', forward direction, needs to be reverse complimented)
pcya PacI buf atgtctcaagttttgtttgatgttattcaataataa ttaattaa gcg
Stats
- Melting: 55.2 C
- Worst Hairpin: -0.46 kcal/mol
- Worst Self-Dimer: -13.61 kcal/mol
Notes
Of course, the issue is that these primers are far from ideal. The pacI site (ttaattaa) allows for very strong homodimers. pacI was chosen however because the normal cloning sites, EcoRI and XbaI, are both found in the actual gene itself, and pacI was one of the few available unique restriction sites that was easy to use / readily available.
pcyA Sequence
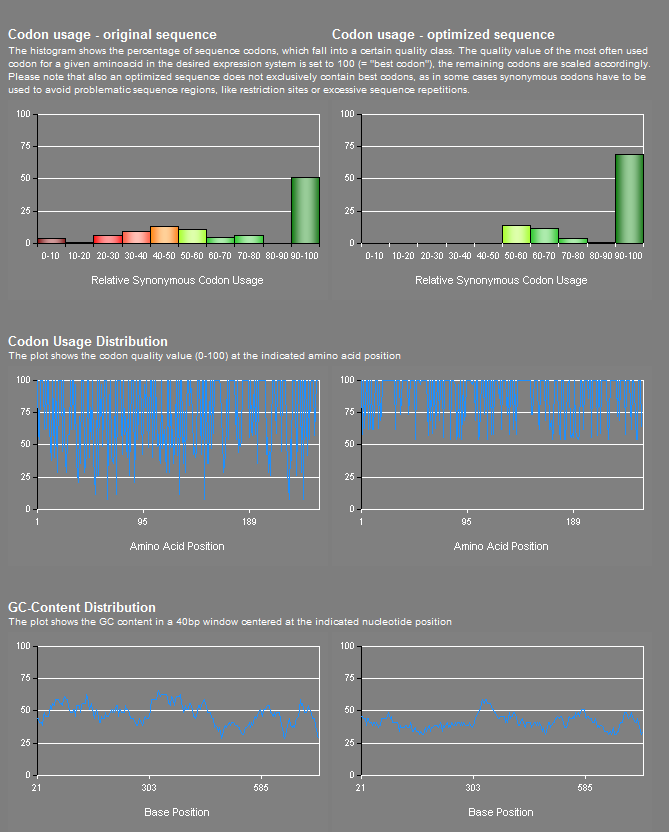
The optimized and unoptimized pcyA sequence. Original sequence is from the registry, part BBa_I15009
Unoptimized
The unoptimized sequence contains 4 instances of a codon that has the absolute lowest expression level in yeast
>BBa_I15009 Part-only sequence (750 bp) atggccgtcactgatttaagtttgaccaattcttccctgatgcctacgttgaacccgatgattcaacagttggccctggcgatcgccgctagttggcaaa gtttacccctcaagccctatcaattgccggaggatttgggctacgtagaaggccgcctggaaggggaaaagttagtgattgaaaatcggtgctaccaaac gccccagtttcgcaaaatgcatttggagttggccaaggtgggcaaagggttggatattctccactgtgtaatgtttcctgagcctttatacggtctacct ttgtttggctgtgacattgtggccggccccggtggagtaagtgcggctattgcggatctatcccccacccaaagcgatcgccaattgcccgcagcgtacc aaaaatcattggcagagctaggccagccagaatttgagcaacaacgggaattgcccccctggggagaaatattttctgaatattgtttattcatccgtcc cagcaatgtcactgaagaagaaagatttgtacaaagggtagtggactttttgcaaattcattgtcaccaatccatcgttgccgaacccttgtctgaagct caaactttggagcaccgtcaggggcaaattcattactgccaacaacaacagaaaaatgataaaacccgtcgggtactggaaaaagcttttggggaagctt gggcggaacggtatatgagccaagtcttatttgatgttatccaataataa
Raw Optimized
Agreed to not use
>Optimized BBa_I15009 (750 bp) ATGGCTGTTACTGATTTGTCTTTGACTAATTCTTCTTTGATGCCAACTTTGAATCCAATG ATTCAACAATTGGCTTTGGCTATTGCTGCTTCTTGGCAATCTTTGCCATTGAAACCATAT CAATTGCCAGAAGATTTGGGTTATGTTGAAGGTAGATTGGAAGGTGAAAAATTGGTTATT GAAAATAGATGTTATCAAACTCCACAATTTAGAAAAATGCATTTGGAATTGGCTAAAGTT GGTAAAGGTTTGGATATTTTGCATTGTGTTATGTTTCCAGAACCATTGTATGGTTTGCCA TTGTTTGGTTGTGATATTGTTGCTGGTCCAGGTGGTGTTTCTGCTGCTATTGCTGATTTG TCTCCAACTCAATCTGATAGACAATTGCCAGCTGCTTATCAAAAATCTTTGGCTGAATTG GGTCAACCAGAATTTGAACAACAAAGAGAATTGCCACCATGGGGTGAAATTTTTTCTGAA TATTGTTTGTTTATTAGACCATCTAATGTTACTGAAGAAGAAAGATTTGTTCAAAGAGTT GTTGATTTTTTGCAAATTCATTGTCATCAATCTATTGTTGCTGAACCATTGTCTGAAGCT CAAACTTTGGAACATAGACAAGGTCAAATTCATTATTGTCAACAACAACAAAAAAATGAT AAAACTAGAAGAGTTTTGGAAAAAGCTTTTGGTGAAGCTTGGGCTGAAAGATATATGTCT CAAGTTTTGTTTGATGTTATTCAATAATAA
Mr. Gene Optimized
Pushed through GeneArt's optimization server
Parameters
Server was asked to avoid:
- EcoRI GAATTC
- Eukaria: (consensus) Splice-Donor (01)
- Eukaria: (consensus) Splice-Donor (02)
- Eukaria: poly(A)-site (01)
- Eukaria: poly(A)-site (02)
- NotI GCGGCCGC
- PacI TTAATTAA
- Prokaria: (consensus) TATA-Box
- Prokaria: -35 Box (01)
- Prokaria: -35 Box (02)
- Prokaria: RBS-Entry (01)
- Prokaria: RBS-Entry (02)
- PstI CTGCAG
- SpeI ACTAGT
- XbaI TCTAGA
- XhoI CTCGAG
- Yeast: poly(A) UE (01)
- Yeast: poly(A) UE (02)
- Yeast: Splice Donor (01)
- Yeast: Splice Donor (02)
Output
>Mr. Gene Optimized BBa_I15009 (750 bp) ATGGCCGTTACCGATTTGAGTTTGACCAATTCCTCCTTGATGCCAACCTTAAACCCTATGATTCAACAATTGGCTTTGGCTATTGCTGCTTCCTGGCAATCTTTGCCTTTG AAACCATATCAATTGCCTGAAGATTTGGGTTATGTCGAAGGTAGATTAGAAGGTGAAAAATTGGTTATCGAAAACAGATGCTATCAAACCCCACAATTCAGAAAAATGCAC TTGGAATTGGCTAAAGTCGGTAAAGGTTTAGACATCTTACACTGTGTCATGTTCCCTGAACCATTGTATGGTTTACCATTATTCGGTTGTGACATCGTTGCTGGTCCTGGT GGTGTCTCTGCTGCCATTGCCGATTTGTCTCCAACACAATCCGATAGACAATTGCCTGCTGCCTATCAAAAATCCTTGGCCGAATTGGGTCAACCAGAATTTGAACAACAA AGAGAATTGCCTCCTTGGGGTGAAATTTTCTCCGAATATTGTTTGTTCATTAGACCATCCAACGTCACCGAAGAAGAAAGATTCGTCCAAAGAGTTGTCGACTTCTTACAA ATCCACTGCCACCAATCCATCGTAGCCGAACCATTATCCGAAGCTCAAACATTGGAACACAGACAAGGTCAAATCCATTATTGCCAACAACAACAAAAAAACGACAAGACT AGAAGAGTTTTGGAAAAGGCTTTCGGTGAAGCTTGGGCCGAAAGATATATGTCCCAAGTTTTATTCGACGTCATTCAATGATGA
DNA 2.0 Quote Optimized
Received back this quote from DNA 2.0
Sequence
>pycA Optimized (DNA2.0) ATGGCTGTGACTGATTTGTCATTGACAAACAGTTCTTTGATGCCAACTCTGAACCCAATGATACAACAGCTTGCACTGGCTATTGCTGCTAGTTGGCAATCTCT ACCTCTTAAACCATACCAATTACCAGAAGATCTGGGTTACGTGGAGGGTAGACTTGAGGGTGAGAAGCTGGTGATTGAGAATAGATGCTATCAAACTCCACAGT TCAGAAAGATGCACTTGGAGTTAGCTAAAGTTGGTAAAGGGTTAGACATCTTACATTGCGTTATGTTCCCTGAACCTTTGTACGGATTGCCATTGTTTGGTTGT GATATTGTAGCAGGACCTGGTGGTGTATCCGCTGCCATTGCAGATCTTTCACCAACTCAGTCTGATCGTCAACTACCAGCTGCCTACCAAAAGTCTTTGGCAGA ATTAGGACAACCAGAGTTCGAACAACAAAGAGAACTGCCACCTTGGGGCGAAATCTTTTCTGAATACTGTTTGTTCATCAGACCATCCAATGTTACCGAGGAAG AAAGGTTCGTCCAAAGAGTCGTTGATTTCTTGCAAATACATTGTCATCAATCTATTGTTGCCGAACCTTTATCTGAAGCACAAACACTAGAACATAGACAGGGC CAAATACACTATTGTCAACAACAGCAGAAAAACGATAAGACAAGAAGAGTACTAGAAAAGGCATTTGGGGAGGCTTGGGCAGAAAGATACATGTCACAAGTCCT ATTTGACGTTATCCAGTAATAA
Raw Output
>Li_NoName_061609_opt
GCGGAATTCGCGGCCGCTTCTAGAGCTCGAGAACATATGGCTGTGACTGATTTGTCATTGACAAACAGTTCTTTGATGCCAACTCTGAACCCAATGATACAACA
GCTTGCACTGGCTATTGCTGCTAGTTGGCAATCTCTACCTCTTAAACCATACCAATTACCAGAAGATCTGGGTTACGTGGAGGGTAGACTTGAGGGTGAGAAGC
TGGTGATTGAGAATAGATGCTATCAAACTCCACAGTTCAGAAAGATGCACTTGGAGTTAGCTAAAGTTGGTAAAGGGTTAGACATCTTACATTGCGTTATGTTC
CCTGAACCTTTGTACGGATTGCCATTGTTTGGTTGTGATATTGTAGCAGGACCTGGTGGTGTATCCGCTGCCATTGCAGATCTTTCACCAACTCAGTCTGATCG
TCAACTACCAGCTGCCTACCAAAAGTCTTTGGCAGAATTAGGACAACCAGAGTTCGAACAACAAAGAGAACTGCCACCTTGGGGCGAAATCTTTTCTGAATACT
GTTTGTTCATCAGACCATCCAATGTTACCGAGGAAGAAAGGTTCGTCCAAAGAGTCGTTGATTTCTTGCAAATACATTGTCATCAATCTATTGTTGCCGAACCT
TTATCTGAAGCACAAACACTAGAACATAGACAGGGCCAAATACACTATTGTCAACAACAGCAGAAAAACGATAAGACAAGAAGAGTACTAGAAAAGGCATTTGG
GGAGGCTTGGGCAGAAAGATACATGTCACAAGTCCTATTTGACGTTATCCAGTAATAATTAATTAATACTAGTAGCGGCCGCTGCAGGCG
>5RE
GCGGAATTCGCGGCCGCTTCTAGAGCTCGAGAACAT
>Li_NoName_061609
ATGGCTGTGACTGATTTGTCATTGACAAACAGTTCTTTGATGCCAACTCTGAACCCAATGATACAACAGCTTGCACTGGCTATTGCTGCTAGTTGGCAATCTCT
ACCTCTTAAACCATACCAATTACCAGAAGATCTGGGTTACGTGGAGGGTAGACTTGAGGGTGAGAAGCTGGTGATTGAGAATAGATGCTATCAAACTCCACAGT
TCAGAAAGATGCACTTGGAGTTAGCTAAAGTTGGTAAAGGGTTAGACATCTTACATTGCGTTATGTTCCCTGAACCTTTGTACGGATTGCCATTGTTTGGTTGT
GATATTGTAGCAGGACCTGGTGGTGTATCCGCTGCCATTGCAGATCTTTCACCAACTCAGTCTGATCGTCAACTACCAGCTGCCTACCAAAAGTCTTTGGCAGA
ATTAGGACAACCAGAGTTCGAACAACAAAGAGAACTGCCACCTTGGGGCGAAATCTTTTCTGAATACTGTTTGTTCATCAGACCATCCAATGTTACCGAGGAAG
AAAGGTTCGTCCAAAGAGTCGTTGATTTCTTGCAAATACATTGTCATCAATCTATTGTTGCCGAACCTTTATCTGAAGCACAAACACTAGAACATAGACAGGGC
CAAATACACTATTGTCAACAACAGCAGAAAAACGATAAGACAAGAAGAGTACTAGAAAAGGCATTTGGGGAGGCTTGGGCAGAAAGATACATGTCACAAGTCCT
ATTTGACGTTATCCAGTAATAA
>3RE
TTAATTAATACTAGTAGCGGCCGCTGCAGGCG
Translation Map
Li_NoName_061609
1 ATGGCTGTGACTGATTTGTCATTGACAAACAGTTCTTTGATGCCAACTCTGAACCCAATG
1 M A V T D L S L T N S S L M P T L N P M
61 ATACAACAGCTTGCACTGGCTATTGCTGCTAGTTGGCAATCTCTACCTCTTAAACCATAC
21 I Q Q L A L A I A A S W Q S L P L K P Y
121 CAATTACCAGAAGATCTGGGTTACGTGGAGGGTAGACTTGAGGGTGAGAAGCTGGTGATT
41 Q L P E D L G Y V E G R L E G E K L V I
181 GAGAATAGATGCTATCAAACTCCACAGTTCAGAAAGATGCACTTGGAGTTAGCTAAAGTT
61 E N R C Y Q T P Q F R K M H L E L A K V
241 GGTAAAGGGTTAGACATCTTACATTGCGTTATGTTCCCTGAACCTTTGTACGGATTGCCA
81 G K G L D I L H C V M F P E P L Y G L P
301 TTGTTTGGTTGTGATATTGTAGCAGGACCTGGTGGTGTATCCGCTGCCATTGCAGATCTT
101 L F G C D I V A G P G G V S A A I A D L
361 TCACCAACTCAGTCTGATCGTCAACTACCAGCTGCCTACCAAAAGTCTTTGGCAGAATTA
121 S P T Q S D R Q L P A A Y Q K S L A E L
421 GGACAACCAGAGTTCGAACAACAAAGAGAACTGCCACCTTGGGGCGAAATCTTTTCTGAA
141 G Q P E F E Q Q R E L P P W G E I F S E
481 TACTGTTTGTTCATCAGACCATCCAATGTTACCGAGGAAGAAAGGTTCGTCCAAAGAGTC
161 Y C L F I R P S N V T E E E R F V Q R V
541 GTTGATTTCTTGCAAATACATTGTCATCAATCTATTGTTGCCGAACCTTTATCTGAAGCA
181 V D F L Q I H C H Q S I V A E P L S E A
601 CAAACACTAGAACATAGACAGGGCCAAATACACTATTGTCAACAACAGCAGAAAAACGAT
201 Q T L E H R Q G Q I H Y C Q Q Q Q K N D
661 AAGACAAGAAGAGTACTAGAAAAGGCATTTGGGGAGGCTTGGGCAGAAAGATACATGTCA
221 K T R R V L E K A F G E A W A E R Y M S
721 CAAGTCCTATTTGACGTTATCCAGTAATAA
241 Q V L F D V I Q * *
Restriction Sites
Name Seq. Locations
AatI AGGCCT none
AccI GTMKAC 187
AflII CTTAAG none
AgeI ACCGGT none
AlwI GGATC none
AlwNI CAGNNNCTG 358
ApaI GGGCCC none
ApaLI GTGCAC none
AscI GGCGCGCC none
AseI ATTAAT 785, 789
AvaI CYCGRG 25
AvaII GGWCC 360
AvrII CCTAGG none
BamHI GGATCC none
BbsI GAAGAC none
BbvI GCAGC 120(c), 378(c), 426(c), 808(c)
BclI TGATCA none
BglI GCCNNNNNGGC none
BglII AGATCT 167, 389
BlpI GCTNAGC none
BsaI GGTCTC none
BsmAI GTCTC none
BsmBI CGTCTC none
BstEII GGTNACC none
BstXI CCANNNNNNTGG none
ClaI ATCGAT none
DraIII CACNNNGTG none
EagI CGGCCG 10, 803
EarI CTCTTC 703(c)
EcoRI GAATTC 3
EcoRV GATATC none
FokI GGATG 535(c)
FseI GGCCGGCC none
HindIII AAGCTT none
KasI GGCGCC none
KpnI GGTACC none
MluI ACGCGT none
NarI GGCGCC none
NcoI CCATGG none
NdeI CATATG 33
NheI GCTAGC none
NotI GCGGCCGC 9, 802
NsiI ATGCAT none
PacI TTAATTAA 786
PciI ACATGT 748
PmeI GTTTAAAC none
PstI CTGCAG 809
PvuI CGATCG none
PvuII CAGCTG 424
SacI GAGCTC 22
SacII CCGCGG none
SalI GTCGAC none
SapI GCTCTTC none
SfiI GGCCNNNNNGGCC none
SgrAI CRCCGGYG none
SmaI CCCGGG none
SpeI ACTAGT 795
SphI GCATGC none
SspI AATATT none
StuI AGGCCT none
SwaI ATTTAAAT none
TliI CTCGAG 25
XbaI TCTAGA 18
XhoI CTCGAG 25
XmaI CCCGGG none
XmnI GAANNNNTTC none
GCRun8 SSSSSSSS 8, 9, 802
T7ClassII YATCTGTW none
W6 WWWWWW 780, 781, 782, 783, 784, 785, 786, 787, 788, 789, 790
SpliceDonor AGGTRAG none
SpliceDonor2 GGTRAGT none
SpliceAcc YYYNYAGGW none
SpliceAcc2 YNCAGGW none
RNADestab ATTTA none
A6 AAAAAA none
C6 CCCCCC none
G6 GGGGGG none
T6 TTTTTT none
ATRich AAWWAA 784, 788, 786(c)
ATRich2 ATATATATA none
PolyA2 AATGAA none
PolyA3 AAATGGAAA none
PolyA4 AATGGAAATG none
SpliceDnr3 GGTAAG none
SpliceAcc3 YYYNCAGRW none
Codon Usage Table
AmAcid Codon Number /1000 Fraction
END TAA 2 8.0 1.0
END TGA 0 0.0 0.0
END TAG 0 0.0 0.0
ALA GCT 8 32.0 0.44
ALA GCA 7 28.0 0.38
ALA GCC 3 12.0 0.16
ALA GCG 0 0.0 0.0
CYS TGT 4 16.0 0.66
CYS TGC 2 8.0 0.33
ASP GAT 7 28.0 0.77
ASP GAC 2 8.0 0.22
GLU GAA 14 56.0 0.63
GLU GAG 8 32.0 0.36
PHE TTC 6 24.0 0.6
PHE TTT 4 16.0 0.4
GLY GGT 7 28.0 0.5
GLY GGA 3 12.0 0.21
GLY GGC 2 8.0 0.14
GLY GGG 2 8.0 0.14
HIS CAT 4 16.0 0.66
HIS CAC 2 8.0 0.33
ILE ATC 4 16.0 0.33
ILE ATT 5 20.0 0.41
ILE ATA 3 12.0 0.25
LYS AAG 5 20.0 0.55
LYS AAA 4 16.0 0.44
LEU TTG 10 40.0 0.33
LEU TTA 6 24.0 0.2
LEU CTA 5 20.0 0.16
LEU CTT 4 16.0 0.13
LEU CTG 5 20.0 0.16
LEU CTC 0 0.0 0.0
MET ATG 6 24.0 1.0
ASN AAC 3 12.0 0.6
ASN AAT 2 8.0 0.4
PRO CCA 11 44.0 0.64
PRO CCT 6 24.0 0.35
PRO CCC 0 0.0 0.0
PRO CCG 0 0.0 0.0
GLN CAA 17 68.0 0.70
GLN CAG 7 28.0 0.29
ARG AGA 10 40.0 0.83
ARG AGG 1 4.0 0.08
ARG CGT 1 4.0 0.08
ARG CGA 0 0.0 0.0
ARG CGC 0 0.0 0.0
ARG CGG 0 0.0 0.0
SER TCT 7 28.0 0.5
SER TCA 3 12.0 0.21
SER TCC 2 8.0 0.14
SER AGT 2 8.0 0.14
SER AGC 0 0.0 0.0
SER TCG 0 0.0 0.0
THR ACA 3 12.0 0.37
THR ACT 4 16.0 0.5
THR ACC 1 4.0 0.12
THR ACG 0 0.0 0.0
VAL GTT 6 24.0 0.4
VAL GTC 3 12.0 0.2
VAL GTA 3 12.0 0.2
VAL GTG 3 12.0 0.2
TRP TGG 3 12.0 1.0
TYR TAC 6 24.0 0.75
TYR TAT 2 8.0 0.25
GC Percentage: 43.39853300733496%
Repeats greater than or equal to 10, in Li_NoName_061609_opt
AGCGGCCGCT (10 bases)
802, 811
802, 811
ATTAATTAAT (10 bases)
786, 795
786, 795
TAATTAATTA (10 bases)
784, 793
784, 793
Cost / Logistics
We have a 2,500 bp limit for the special rates of $0.2/bp.
The construct itself is 843bp. At 0.2$/bp, it is roughly 170 dollars. Plus $35 for shipping, total ends up at $204
Turnaround is ~ 15 days.
This also leaves us with roughly 1657bp left for additional synthesis at the special iGEM price.