IGEM:MIT/2007/Notebook/2007-7-19
From OpenWetWare
Jump to navigationJump to search
Agenda for Thursday, 7/19
- Make permanent glycerol stocks of 8 tubes
- AK/AL list
- Colony PCR on yesterday's plated transformation (F2620+B0034+CPX in pSB1AC3)
Colony Count of Yesterday's Plates
TK's DH5a transformed with F2620+B0034+CPX in pSB1AC3
Key:
- (+) contains DH5a transformed with digested and ligated parts
- (-) contains DH5a transformed with only digested and ligated pSB1AC3
- BH the plate is labeled with Bernice's initials
- JH the plate is labeled with Jess's initials
Plate #Colonies Notes (+) Cm BH ~50 A couple satellite colonies (+) Cm + Kan BH 1 This plate hardened imperfectly (+) Cm JH ~64 (+) Cm + Kan JH 3 On the lip of the plate (-) Cm 9 (-) Cm + Kan 0
Colony PCR of Yesterday's 3A Transformation
- Used 8 colonies picked from (+) Cm + Kan JK plate
- Streaked those 8 colonies also onto Cm and Cm + Kan plates for antibiotic screening
- Used the OWW Knight Protocol for Colony PCR (see below)
Colony PCR Protocol (OWW, Knight)
- Prepare one sterile 0.6mL tube with 20 µl ddH20 for each colony
- Prepare LB-agar plate with appropriate antibiotic to use as index plate
- Pick single colony using a pipetman with sterile tip. The pipettor should be set to 3µl
- Innoculate tip with colony into tube. Pipet up and down to ensure cells are transferred to tube. Pipet 3µl of cells suspended in water onto index plate.
- Repeat for as many colonies as you intend to pick
Reaction Mixture:
- 9µl PCR supermix
- 0.25 µl 40µM VF2
- 0.25 µl 40µM VR
- 0.5 µl colony template
PCR conditions:
- 95C for 15 mins
- 94C for 30secs
- 56C for 30secs
- 68C for 1 min per kb of expected product
- Repeat 2-4 39 times
- 68C for 20 mins
- 4C forever
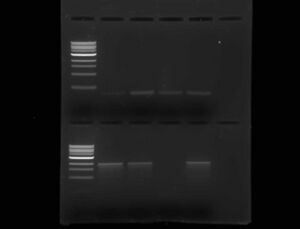
- Run a gel to determine amplification product length
- 5 mins @ 40V, remainder @ 85V
- Used 50ml gel with two rows of 6-wells each
- Row 1, well 1: 1kb ladder
- Row 1, wells 2-5: Colony PCR samples #1-4
- Row 2, well 1: 1kb ladder
- Row 2, wells 2-5: Colony PCR samples #5-8
Permanent Glycerol Stocks of DB3.1, F2620, B0034 and E1010
- SK: Made eight cryogenic vials of glycerol stock; all have 1 mL 40% glycerol + 1 mL liquid sample
- All vials in the white sticker labeled "iGEM 2007 DH5alpha" -80 degree freezer box
- DB3.1, 2 vials = original with pSB3K3 (K+)
- F2620, 2 vials = transformed (A+)
- B0034, 2 vials = transformed (A+)
- E1010, 2 vials = transformed (K+)
Liquid Culture of BBa_I13500
- Last night's growth was unsuccessful, so it was redone
Made More Plates
- just LB: 10 plates
- Cm+Amp: 30 plates
- Cm: 50 µg/ml
- Amp: 30 µg/ml
Protocol for de-phosphorylating DNA before ligating
- Add 1 ul Calf Intestinal Phosphatase or Alkaline Phosphatase
- 37 C for an hour
- Heat Kill
- Purify on Gel