IGEM:MIT/2006/Notebook/2006-6-29
PCR cleanup / Nanodrop / ran gel - TAKE 3
- PCR cleanup: followed standard protocol for PCR cleanup...not too exciting : ) Just remember to elute with 30μL of the last buffer.
- Nanodrop: check the concentrations of the cleaned-up cut DNA:
- Plasmid (pSB1A3-1, cut with EcoRI and PstI): 29.8 ng/μL
- Barry's part (PSBI82E0040, cut with EcoRI and PstI): 11.7 ng/μL
- BSMT (cut with EcoRI, PstI, and DpnI): 12 ng/μL
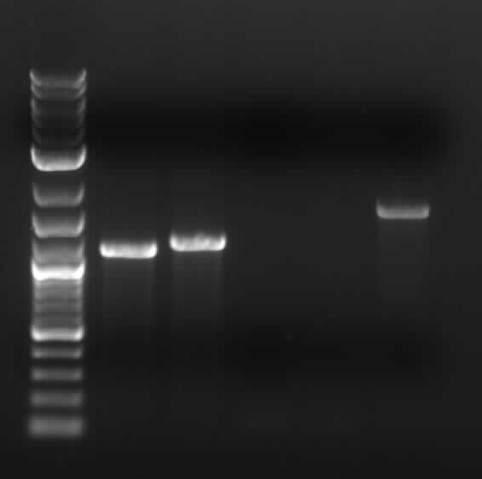
- Gel (The 3 tubes of cut DNA are in the yellow holder...use the set labeled 6/28): the columns, from left to right, are:
- 2log ladder
- cut BSMT (100ng) ... expected length = 1.1kB
- cut Barry's biobrick part (100ng) ... expected length = 2kB backbone + .72kB fragment
- cut pSB1A3-1 backbone ... expected length = 2.1kB + another short fragment
- uncut pSB1A3-1 backbone (2μL, aka 200ng)... expected to look crazy b/c it's a circle
Update on Stationary Phase Promoter
Using Francois's promoter attached to a fluorescent protein on a plasmid, we made a liquid culture and a glycerol. However, the liquid culture of his control plasmid did not grow. Before we can test the fluorescent rate of the promoter over time, we need to get a liquid culture treated with the control plasmid to grow. Two more plates are being grown with colonies containing this plasmid and another liquid culture is in the process of being made from one of the colonies from the original plate. I am waiting to talk to Tom about our options regarding how we should go about obtaining osmY, which is said to be a stronger stationary phase promoter than Francoiss.
Smell test Results
The proper liquid cultures smelled!
GC Training
Stephen and Veena were trained to use the GCMS. We received the proper syringes to operate the machine, and we now have cultures to investigate on the machine.
Ligation (BECAUSE DIGEST FINALLY WORKED!!!)
We'll do 2 ligations: one with the cut pSB1A3-1 backbone and our cut BSMT DNA, and another one with the cut pSB1A3-1 backbone and Barry's cut biobrick part.
General ligation considerations: 10μL total volume, about 50ng of each DNA
BSMT Ligation
- 1.5μL water
- 1μL T4 DNA ligase buffer (vortex well before adding, make sure it smells strongly like wet dog!!)
- .5μL T4 DNA ligase enzyme
- 5μL cut BSMT (50ng)
- 2μL cut backbone (50ng)
Control Ligation (Barry's biobrick part)
- 1.5μL water
- 1μL T4 DNA ligase buffer (vortex well before adding, make sure it smells strongly like wet dog!!)
- .5μL T4 DNA ligase enzyme
- 5μL cut Barry's BB part (50ng)
- 2μL cut backbone (50ng)
Ligate at room temp for 15 to 20 minutes and then proceed to transformation
PCR with new SAMT, ATF1, and JMT primers
- Doublecheck that SAMT and BSMT genome are dilluted to 1ng/μL in water, and don't mess with yeast genome concentration b/c we know that 1 μL works well
- Prepare 25μM tubes of each primer from our invitrogen order immediately
- In each tube we want a total reaction volume of 50μL, with 1ng of template DNA, .6μL of each primer (forward and reverse), and 49 μL of Tom's PCR mix.
- NOTE---.6 μL of the 25μM primer yields roughly 300 nM final concentration of that primer
- 5 PCR Tubes: BSMT (w/ both BSMT primers), SAMT (w/ both SAMT primers), A. thalinia genome (w/ both JMT primers), A. thalinia ZapII cDNA library (w/ both JMT primers), yeast genome (w/ATF1 primers)
- Run the tubes on the thermocycler at: 95deg for 3:00, then cycle through: (a) 94deg for :30 (b) 55deg for :30 (c)68deg for 2:15, then 72deg for 10:00
- This is the gel, with the lanes (from left to right) being: (1) 2 log ladder (2)BSMT (3)SAMT (4)JMT ZapII (5)JMT (6)ATF1
- So, it looks like the SAMT and the ATF1 worked with the new primers! yay!
- Nanodrop of the cleaned-up PCR products: SAMT = 75.5ng/μL, ATF1 = 30.8ng/μL, BSMT = 64ng/μL
Yeast
- Use as biosensor
Transformation of BSMT biobrick (and controls)
- Thaw Tom's new Top10 cells (don't use ones that have been thawed and refrozen)...then proceed with the transformation like we normally do
- 30 min incubation on ice, SOC is critical, 50 sec heat shock!!, 1 hr incubation with rotation in 37c room
- label 6 Amp plates and plate both 20 μL and 200 μL on these only Amp plates of BSMT ligation, Barry's Part ligation, and pUC19 --- yay positive controls!
- incubate plates overnight in 37c room