IGEM:IMPERIAL/2007/Tutorials/Guide for Engineers/Central Dogma of Molecular Biology
Note: Images used are not copyrighted. A big thanks to Dr. Steve Cook (Imperial College) for permitting the use of his images and lecture notes. Other images obtained from Wikipedia.
Introduction
“The central dogma of molecular biology deals with the detailed residue-by-residue transfer of sequential information. It states that such information cannot be transferred back from protein to either protein or nucleic acid.” - Francis Crick, 1958
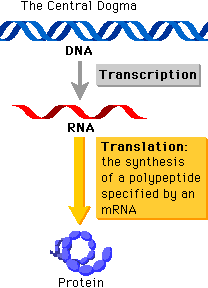
It might seem like a whole load of jargon, but the basic concept underlying the central dogma is that the flow of information proceeds in a linear step-wise fashion, from DNA to RNA and finally protein. This means that protein information cannot flow back to that of nucleic acids. The central dogma is a fundamental concept that basically forms the structural framework with which molecular biologists understand the transfer of sequence information between the different biopolymers.
So why is the central dogma important to synthetic biology? Many recombinant DNA technologies, as with our understanding of cellular processes, are more often than not based or adapted from what we know. Knowledge is power, and the more we understand, the more we are able to manipulate its machinery to meet our required specifications. And we do know a lot about the mechanisms behind it.
The Basics: Cell Organization and DNA Structure
The cell is a compartment (not necessarily the simplest form of a living system), and has the ability to localize concentration of metabolites, substrates, and catalysts. Cells are always bounded by an amphipathic lipid bilayer, usually made of phospholipids.
General similarities:
- Proteins: L-amino acids (common chirality – D-amino acids are very rare – peptidoglycan).
- Carbohydrates: glucose/glycolysis as main energy source, α-(1→4) glucans for energy storage, β-(1→4) glucans (cellulose, chitin) for structural purposes.
- RNA: universality of the genetic code, all cells possess very similar ribosomes.
- DNA: almost invariably the genetic material.
- Lipids: terpenes and phospholipids used for membranes.
- Porphyrins: haem, chlorophyll, bilin pigments, all with tetrapyrrole motif. Used for redox (electrons, oxygen, etc.).
Prokaryotes
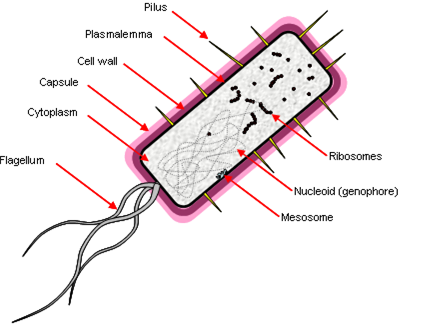
Prokaryotes have a simple cell morphology, and are characterized by the lack of a nucleus, leaving a free circular genophore.
Other properties:
- ‘Simple’ rotating motor-type flagellum.
- Cell division by fission, genophore attached to plasmalemma by mesosome.
- Few membrane-bound organelles, no double-membrane bound organelles.
- They are ‘small’ because diffusion limits the rate of transport across the cell – 1 μm.
Prokaryotic DNA also lacks histones (packaging proteins) and molecules have direct access to DNA. They have a junk-free genome, transcribed by just one RNA polymerase. Transcription leads to translation with no intermediate processing, which allows several genes to be encoded in a single transcriptional unit (a polycistron). They have small (70S) ribosomes.
Eukaryotes
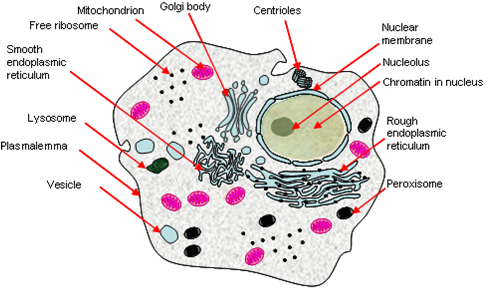
Eukaryotes have a complex system of internal organelles (although not necessarily more complex by nature), and is characterized by the presence of a nucleus with its genophore enclosed within.
Other properties:
- Double-membrane bound nucleus containing linear chromosomes.
- Complex 9+2-type undulipodium, cytoskeleton, cytosis and mitosis.
- Many membrane-bound organelles and double membrane-bound endosymbionts (e.g. mitochondria, chloroplasts).
- They can grow ‘large’ because cytoplasmic streaming allows rapid transport across the cell – 100 μm.
Eukaryotic DNA is bound by histones, and requires some degree of unpacking for expression. Much of their genome is composed of parasitic DNA and introns. Three RNA polymerases exists, (approximately) one for each sort of major RNA product:
- RNApol-I – rRNA.
- RNApol-II – mRNA and snRNA.
- RNApol-III – tRNA and 5S rRNA.
RNA is heavily processed in the nucleus (e.g. splicing mechanisms). They possess larger 80S ribosomes.
Structure of DNA
Nucleic acids may occur in double or single stranded forms, most typically:
- double stranded DNA (dsDNA)
- single stranded RNA (ssRNA)
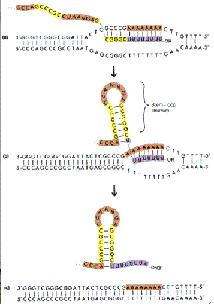
Nucleic acids can also pair with themselves to form hairpin loops, cruciforms, internal loops and bulges. These are important in the termination of transcription and restriction enzymes (palindromic sequences pair readily with themselves). Secondary and tertiary structure of RNAs can give it catalytic activity.
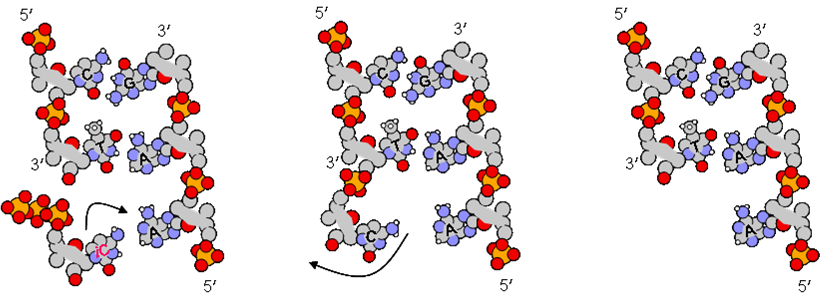
DNA consists of two nucleotide polymers that form a right-handed double helix, with a backbone made of ribose sugar and phosphate groups joined by phosphodiester bonds. Attached to each ribose is one of four bases - adenine (A), thymine (T), cytosine (C), and guanine (G). Essentially the sequence of these four bases form that basis of the genetic code, where information is passed along the central dogma, as to the next generation. At this point we should also note that RNA is encoded by uracil (U) as opposed to thymine.
Within cells, DNA is organized into structures called chromosomes, and a set of chromosomes within the cell make up the genome.
For prokaryotes, this is largely a simple loop of dsDNA, although some viral and other parasitic ssDNA, ssRNA, dsRNA, etc., may be present. The majority of the genome is present on a single chromosome - in Escherichia coli this is about 5 megabases (Mb) in size, and supercoiled.
For eukaryotes, the genome is found in the nucleus, where chromatin proteins like histones bind to it to compact it further, and also control its interactions with other proteins and therefore transcription rates.
In addition to genomic DNA, extrachromosomal DNA are also present in the form of other organelles like mitochondria and chloroplasts. Other examples include plasmids, an important vector in recombinant DNA technology, and bacterial artificial chromosomes (BACs) and yeast artificial chromosomes (YACs).
Gene Organization
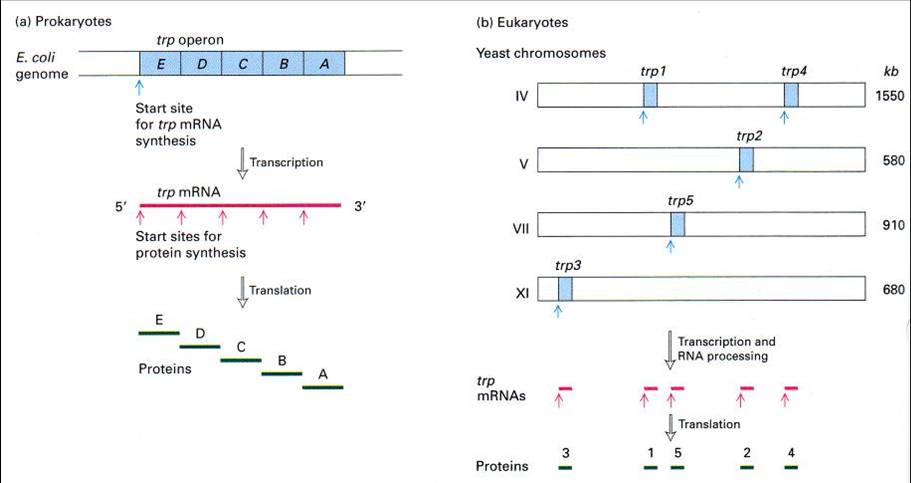
The way that genes are organized in Prokaryotes are also significantly different from that of Eukaryotes. In Prokaryotes, genes are usually organized in operons, where a single operon is responsible for the synthesis and regulation of proteins that are associated with its function. A polycistron (cistron being the region where mRNA is translated) is also formed which eventually translates to the various proteins within the operon.
In contrast, Eukaryotes often have genes in discrete locations, and its mRNA is monocistronic. Due to the difference in gene organization, its genes are also regulated in different ways. Refer to the lac operon section for more details.
DNA Replication
DNA replication is the process of copying double-stranded DNA molecules.
What must replication achieve?
- DNA is a helix.
- Must be unwound and ssDNA must be stopped from annealing to inappropriately.
- DNA is a very long, twisted molecule.
- Must avoid tangling.
- DNA is information.
- Must be copied with high fidelity.
- DNA is a double helix.
- Must synthesise two strands simultaneously in 5ʹ→3ʹ and 3ʹ→5ʹ directions.
This is achieved by various proteins as will be explained later. Two important concepts are associated with DNA replication:
- Semi-conservative replication
- Proofreading mechanisms
Semi-conservative Replication
http://upload.wikimedia.org/wikipedia/en/a/a2/DNAreplicationModes.png
Theoretically, the copying of DNA can be achieved via several means. Semi-conservative replication is the only known method to be used in Nature.
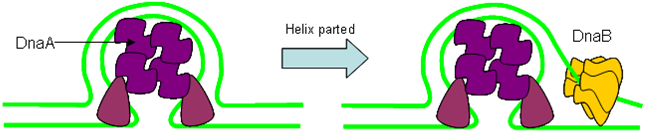
Origin of Replication
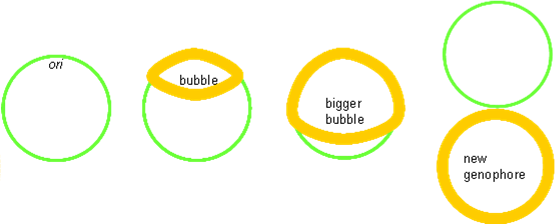
The origin of replication is a length of DNA into which proteins can insert and form a ‘replication bubble’ by loading in DNA polymerase. Two replication forks migrate from the bubble outwards, synthesising DNA as they go.
Prokaryotic chromosomes only have one origin of replication (ori), and its entire genome (5 Mbs) are copied in about 40 min, which means a speed of ≈ 1000 bp s−1. Its rate of growth is further increased in complete media.
From this on we would omit anything that is to do with eukaryotes unless otherwise stated.
Enymes involved
As stated above, there are several criteria for copying DNA so as to achieve fecundity, fidelity and longevity. This is achieved by various proteins that alter its shape (topology). The melting point of strands is c. 90°C in vitro (see PCR); this must be made to happen at c. 37°C in vivo.
- Initiator Proteins
Initiator proteins involved in 'melting' of DNA helix at ori to allow loading of helicase.
- Helicase
Helicase involved in energy-dependent separation of DNA strands by screwing mechanism.
- Single stranded binding proteins (SSBPs)
SSBPs stabilise the parted strands, which would otherwise anneal to themselves (or other strands), forming hairpin loops and other unwanted tangles.
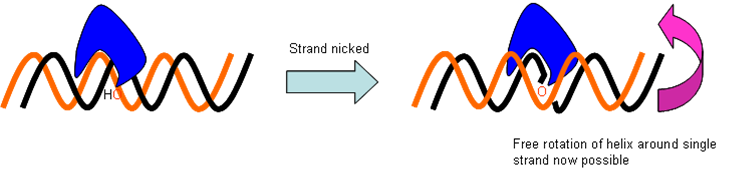
- Topoisomerase I
Topoisomerases stop tangles. Topoisomerase I nicks the strand ahead of helicase, allowing strain-relief, and preventing snarling up of the strands as they are forced apart.
- Topoisomerase II
Prokaryotic chromosomes are Möbius strips. Topoisomerase II creates gates, allowing helices to cross one another. Tangles and linked loops can therefore be separated.
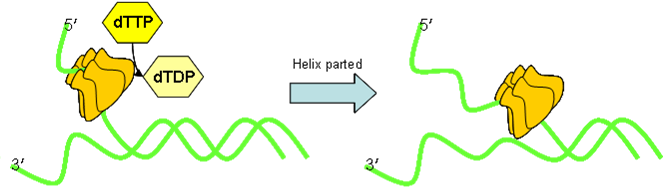
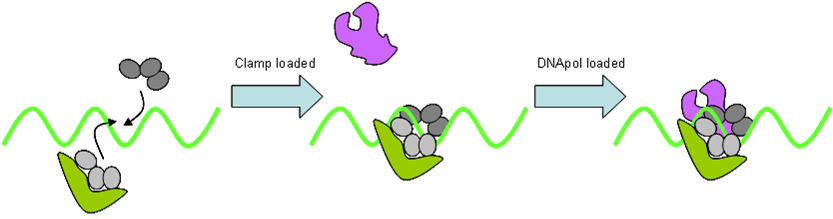
- DNA Polymerase
DNA polymerase is held onto the DNA by a doughnut shaped sliding clamp (processivity factor). This is loaded onto the DNA by a clamp loader. DNA is synthesised from dNTPs. Hydrolysis of (two) phosphate bonds in dNTP drives this reduction in entropy. This however only occurs in the 5ʹ→3ʹ direction. Prokaryotes possess three polymerases:
- DNApol I – repair
- DNApol II – cleans up Okazaki fragments
- DNApol III – main polymerase\
- DNA Primase
DNApol will not synthesise dsDNA from ssDNA: it needs a short double stranded section. RNA primers are therefore added by DNA primase. The primase plus the helicase are often called the lagging strand primosome.
Priming is the most likely point for mismatch to occur because the short helix is unstable, so DNApol has been evolutionarily tuned to refuse to bind to ssDNA, and will only bind to dsDNA or DNA/RNA hybrid. RNA is distinguishable from DNA and can be removed later, whereas a DNA primer would not allow this erasibility. Hence RNA is used.
- DNA Ligase
DNA ligase combine strands of DNA together by linking the phosphate groups on the ribose sugar to form a phosphodiester bond.
The Replicosome
The leading strand is synthesised continuously 5ʹ→3ʹ. However, the other, ‘lagging’ strand is still synthesised 5ʹ→3ʹ but in discrete chunks, from the replication fork back towards the origin.
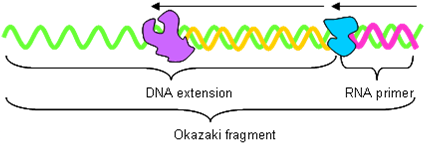
Okazaki Fragments
DNApol produces ‘Okazaki’ fragments on the lagging strand.
- DNA primase adds 10 bp RNA primer to DNA.
- DNApol extends the primer to 200 bp (eukaryotes – nucleosome size?) or 1000 bp (prokaryotes) with DNA.
DNApol drops off when it hits a primer on lagging strand. The RNAse-H complex removes the primer and DNApol fills in the hole. Finally, DNA ligase joins the fragments together. This palaver also needs to be done when DNApol completes the full-circle in prokaryotes.
Proofreading Mechanisms
Binding of nucleotides has error of c. 10−4, due to extremely short-lived imino and enol tautomery. However, the lesion rate in DNA is only 10−9. This increased accuracy is due to the fact that DNApol can chew back mismatched pairs to a clean 3ʹ end using its built-in 3ʹ→5ʹ exonuclease activity.
Imino-cytosine pairs (incorrectly, as far as the cell is concerned) with adenine. Nanoseconds later, the iC converts back to the normal amino form of cytosine, and no longer pairs with adenine. The mismatched base sticks out and is cleaved off by the exonuclease activity of the enzyme, leaving a fresh OH group to try again.
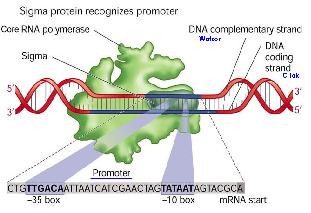
Transcription
t is the process of copying DNA to RNA.
It differs from DNA synthesis in that only one strand of DNA, the template strand, is used to make mRNA.
Unlike DNA replication, it does not need a primer to start.
It can involve multiple RNA polymerases.
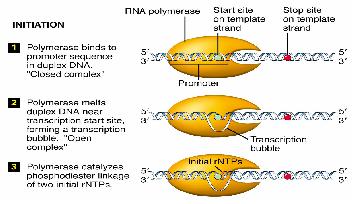
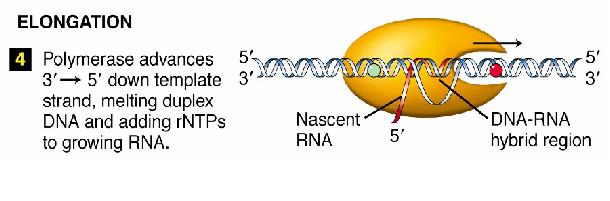
Transcription is divided into 3 stages
- Initiation
- Elongation
- Termination
Initiation
RNA polymerases execute specificity by having a sigma factor, and also an α subunit that recognises a specific region of DNA upstream of the promoter.
Elongation
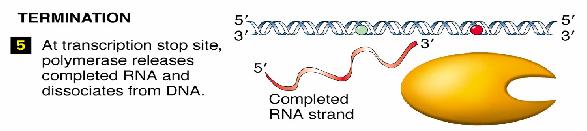
Termination
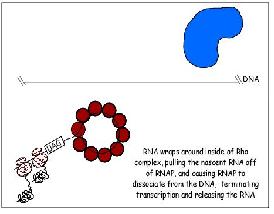
Two types of termination exists, a rho independent type and a rho dependant type. The rho independent type that relies on the weak base pairing that exists between the long continuous chain of A-U base pairing, which results in a particularly flexible portion of the DNA. This can cause the RNA polymerase to dissociate from the DNA and completed mRNA by formation of a stem loop that disrupts the smooth processing of the RNAP along the DNA.
The other type of termination, the rho dependant type, involves a rho protein that yanks the DNA and RNAP apart when they are bound too tightly together and cannot dissociate.
Final Product
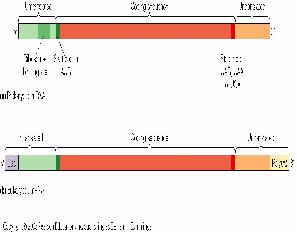
The final product of transcription of a prokaryote gene includes a portion for gene translation, as well as regions at the beginning and the end of the gene that does not get translated.
Transcriptional Control
There are different promoters for different sigma factors. A sigma factor has to bind to a promoter as well as a RNAP to enable the RNAP to start transcribing the gene. It provides specificity for the RNAP as an RNAP is able to bind to any promoter sequence, and thus transcribe any gene. However, as only certain genes need to be transcribed, sigma factors are produced by the cells specific for the promoters of the transcribed genes.
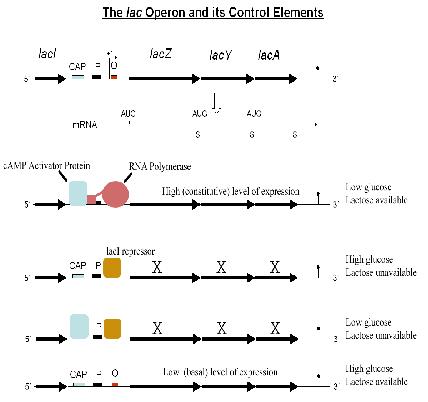
Case Study: Lac Operon
- For control of lactose metabolism
- Consists of three structural genes, a promoter, a terminator and an operator
- LacZ codes for a lactose cleavage enzyme
- LacY codes for ß-galactosidase permease
- LacA codes for thiogalactoside transcyclase
- When lactose is unavailable as a carbon source, the lac operon is not transcribed.
The regulatory response requires the lactose repressor. The lacI gene encoding repressor lies nearby the lac operon and it is consitutively (i.e. always) expressed. In the absence of lactose, the repressor binds very tightly to a short DNA sequence just downstream of the promoter near the beginning of lacZ called the lac operator. Repressor bound to the operator interferes with binding of RNAP to the promoter, and therefore mRNA encoding LacZ and LacY is only made at very low levels. In the presence of lactose, a lactose metabolite called allolactose binds to the repressor, causing a change in its shape. The repressor is unable to bind to the operator, allowing RNAP to transcribe the lac genes and thereby leading to high levels of the encoded proteins. More details can be found here: [[1]]
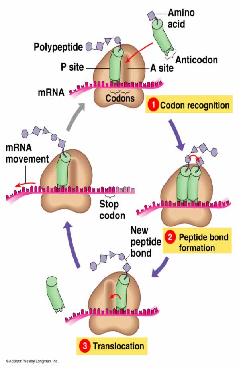
Translation
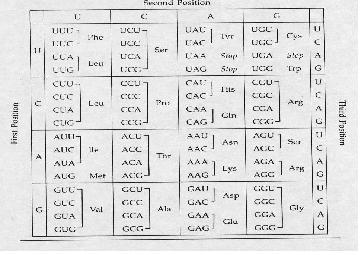
- Interpreting the information coded in the mRNA into proteins.
- The nucleotides are read in triplets (set of three) called codons.
- Each triplet code for a specific amino acid, and sometimes more than one codon exist for an amino acid.
- mRNA are read by the translational machinery including ribosomes, tRNAs and rRNAs.
- Like transcription, it also includes initiation, elongation and termination.
Codon Table
It is important to take note of the STOP codons, which are UAA, UGA, UAG; as well as the start codon AUG which codes for methione. In prokaryotes, the first AUG always codes for formyl methione; while in eukaryotes, the first AUG codes for methione, like all the other AUGs in the rest of the mRNA.
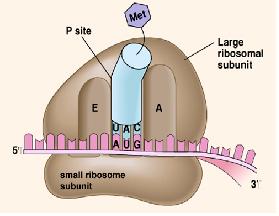
Ribosome
The ribosome is the main machinery for translation. It consists of two subunits, the larger one and the smaller one. It is made of ribosomal RNAs (rRNAs) and proteins. It also has three sites, the A (amino) site, the P (peptidyl) site, and the E (exit) site.
tRNA the Middle Man
- It is in a clover shaped structure.
- It serves the function of bringing the amino acids to the mRNA.
- It has an anticodon loop to recognise the codons in the mRNA (by Watson-Crick base pairing).
- It is responsible for the specificity of the codon recognition.
tRNA Charging
Aminoacylation is the process of adding an aminoacyl group to a compound.
tRNA is aminoacylated(or charged) with a specific amino acid by an aminoacyl tRNA synthase.
There is normally a single aminoacyl tRNA synthetase for each amino acid, despite the fact that there can be more than one tRNA, and more than one anticodon, for an amino acid.
Initiation
In bacteria, polycistronic messages (which have several start codons) have Shine-Dalgarno sequences c. 5 nt to 5ʹ of the AUG start codons. This binds the anti-Shine Dalgarno sequence at the end of the 16S SSU rRNA, ensures that the ribosome binds at the correct place(s), not at an out-of-phase codon or an internal methionine.
mRNA 5ʹ…–AGGAGG–––––AUG–…3ʹ
rRNA 3ʹ–auUCCUCCacuag–…5ʹ
Initiation requites the coming together of the tRNA-fmet, ribosome, and the mRNA to form the full functional machinery for translation.
Elongation
Peptide bond formation is performed by peptidyl transferase, whose active site contains an adenine ring from the 28S rRNA in large subunit. This performs and acid/base catalysis, just like histidine does in many enzymes.
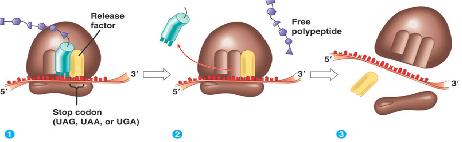
Termination
Stop codons are bound by cytoplasmic release factors, which add water to tRNApeptidyl cleaving off the polypeptide.
Protein Trafficking and Cell-cell Communications
Lipid Bilayer
Most biological molecules are impermeable to the lipid bilayer based on size and charge. Exception applies to very small uncharged molecules (eg. water, carbon dioxide, nitrogen), or small signaling molecules such as acyl homoserine lactone (AHL) in quorum sensing and steroid hormones in humans.
Protein Trafficking
Channels are required to transport molecules across the membrane, and are usually specific to a certain molecule - sometimes even specific to the direction of transport (only import or export). Some channels transport two molecules together, using the potential of one molecule to assist the transport of the second molecule.
Channels can also be classified as passive (allows diffusion only) or active (uses energy to transport against the concentration gradient), and must also be able to open and close to control the flux.
Proteins are usually very large molecules and thus cannot diffuse through the lipid bilayer or use channels. Proteins translated in the cells are usually not folded prior to export, and signal sequences are added as extra bits of translated protein that tags it for export. The membrane transporters then recognize these tags and transport tagged proteins out of the cell.
Quorum Sensing
Quorum sensing is a type of autocrine signaling, and is used by Vibrio fischeri to release light at high cell counts.
http://parts.mit.edu/registry/images/thumb/b/bc/Luxrreceiverschematic.png/800px-Luxrreceiverschematic.png
Source: Registry of Standard Biological Parts
The lux operon is regulated by 2 proteins:
- luxR (lux regulator gene) is transcribed constitutively to produce LuxR. LuxR can only stimulate translation of luxpR (lux promoter) in the presence of molecule acyl homoserine lactone (AHL).
- luxI (lux inducer gene) is transcribed at basal (low) levels, producing low levels of LuxI. LuxI, is an enzyme that converts S-adenosylmethionine (SAM) into AHL which then diffuse s across the cell membrane. It is stable in growth media over a range of pH.
Low Cell Density
At low AHL concentration, there are low levels of LuxR-AHL complex formation, and thus low levels of transcription of luxpR.
High Cell Density
At high AHL concentration, more cells are able to produce LuxI and AHL, thereby leading to more LuxR-AHL complex formed. This allows increased activation of luxpR, leading to a higher transcription rate of luxI. As more LuxI is generated, it acts in a positive feedback to the system to generate more AHL.
Downstream genes (luxCDABE) code of luciferase (an actuator) which generates light.
Therefore, at high cell density, light is generated.
Protein Switches
Transcriptional control of proteins is slow (gene has to be transcribed, translated; protein has to be folded…). Proteins can be modified after translation to activate or deactivate it, the most common modification being the addition of a phosphate group from ATP.
- Kinases add phosphate groups to proteins (requires ATP)
- Phosphatases remove phosphate groups from proteins
The state of phosphorylation thus determines whether the protein is active or inactive.
Membrane Receptors
In the case where large signaling molecules cannot diffuse through the lipid bilayer, surface receptors that recognize the signaling molecule are still able to transduce the signal to achieve its required effect. Receptor tyrosine kinases (RTK) are the most common type of receptor in bacteria:
- The signaling molecule brings two receptor molecules together, allowing cross-phosphorylation
- One receptor acts as a kinase which adds a phosphate group to the other receptor
- Activated receptors can then activate cytoplasmic proteins, achieving downstream signaling pathways which ultimately affect transcription rates.
Two-Component System
The two-component system is also another signal transduction pathway. An external signal can activate the sensory domain of the membrane receptor (found on the extracellular side), causing autophosphorylation of the kinase domain that is found on the intracellular side of the receptor. The kinase domain then transfers phosphate groups to response regulator in the cytoplasm, and activated response regulator can then proceed to turn on expression of specific genes.
Criticisms of Central Dogma and Alternative Thinking
The reductionist approach of the central dogma is what many scientists in systems biology complain about. Indeed we find many evidential circumstances in Nature:
- Reverse transcriptase allows DNA → RNA (HIV, retrotransposons)
- Some RNA viruses never have a DNA form and are perfectly capable of RNA replication.
- Prions (CJD, BSE) are ‘self-replicating’ proteins.
Contemporary thought seems to regard these evidences not as exceptions to the central dogma, but rather that the central dogma is too simplistic in its approach to understanding information transfer in living systems. Indeed to use the central dogma as a research strategy would simply inhibit novel approaches to understanding more complex (or diverse) systems. Therefore a revised version of this would be:
http://upload.wikimedia.org/wikipedia/en/thumb/4/46/CDMB2.png/599px-CDMB2.png
In addition, in vitro gene technology has also allowed for reverse engineering, and the use of proteins to generate the codon sequence to which we could obtain and synthesize the DNA sequence - a method that is commonly used to determine the specific gene location of a protein.