IGEM:IMPERIAL/2007/Projects/Biofilm Production/Notes
Biofilm Production: Notes
Basic Technology
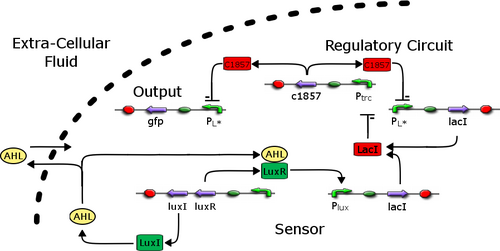
- The core technology of our device has already been developed by Boston Unversity in 2004.13
- The toggle switch is a bistable system for which transitions can be induced by degrading or introducing antagonistic molecules.
- Switching between the high lambdaCI and high LacR regions is brought about by increasing the lambdaCI decay rate or the LacR production rate.(lambdaCI codes for lambdaci , lacI codes for LacR)
- In our system expression of lacI gene is based on the quorum sensing pathway V.fischeri, in short the greather the concentration of AHL molecules the grater the expression of LuxR. This process is illustrated to the right.
Potential Targets For Regulation
We have looked through a variety of literature concerning biofilms and have identified three possible genetic targets to regulate in order to control biofilm formation:
- The genes required for formation of proteins involved in the initial attachment of microorganisms to a surface.
- The genes required for the formation of the exopolysacchorides that are secreted from the microorganisms.
- The genes that are involved in the regulation of these two systems. These really are the regulatory pathways involved.
After looking at both various points of regulation we looked in more detail at specific genes and in particular any knock out studies that have shown their affect on biofilm formation.
Regulatory pathways
Two component systems are key for biofilm formation. There are a variety of examples, three which seemed to be important are:
- BarA/UvrY
- Cpx
- EnvZ/OmpR
We looked at the BarA/UvrY in detail and this is described below.
BarA/uvrY Pathway:
BarA/uvrY are a two component system found in E.coli cells. The BarA and uvrY are involved in the regulation of csrA (carbon storage regulator). CsrA is a repressor of biofilm formation, because it is a RNA binding protein that binds to the mRNA of PSG. PSG is a key molecule that is involved in three processes;
- solid surface attachments.
- cell-cell adhesion.
- stabilisation of biofilm.
The role of BarA and uvrY in the regulation of csrA is that it is involed in the autoregulation of CsrA, that is to say that CsrA autoregulates itself through this two component system. The way that CsrA does this is to activate the transcription of a CsrB RNA. Looking at the literature on this area we have identifies several possible genes that we could try to control.
- CsrA could be regulated under a inducible promoter, however, if this was the case we need to consider the activation of csrB by the BarA/urvY pathway.
- Try to interact with the autoregulation of CrsA
However, the CrsA also controls several other key processes in the cell, including;
- glycogen biosynthesis, catabolism and gluconeogenosis,
Interfering with these pathways could have more adverse affects than biofilms. However, a study done on K12 mutants showed that the only affects of knocking out the BarA/UrvY is to increase the sensitivity to hydrogen peroxide and to alter the carbon utilization response of the E.coli.
wcaF9
In a studied based on K-12 strains of E.Coli the gene wcaF needs to be expressed for the mature development (3D) of Biofilm. The wcaF9 is not involved in the initial binding of the E.Coli to the surface but appears to be important after the binding of the surface. The fact that the expression of wcaF is induced by binding of a surface also inicated this. A recent study compared a strain where the wcaF (wcaF-1) had been knocked out to a wild-type E.coli (wcaF+1) . It was found that the mutant had decreased formation of biofilms. It was only after a given time ~100hours, that the biofilm reached the same level as when wcaF was present. What was clear was that in the wcaF-1 the typical architecture of the wild types biofilm was not present and that as a result the biofilms were less structurally stable. We propose to use this wcaF gene in a wcaF-1 E.coli strain to allow regulation of the biofilm formation. Using this wcaF we will not be able to control the intial attachment, but will be able to control the speed and the architecture at which the biofilm forms.
Control of Two-Component Systems
A reaccuring problem with trying to control the formation of biofilms with the two-component systems is that biofilm formation is only one of many affects downstream of these systems. So, by regulating these two-component systems we will be affecting a greater diversity of cellular function. For this reason we decided to pursue the exopolysacchoride secretions.
Exopolysacchoride production
One of the key components of exopolysacchorides is the production of colanic acid. We thought that if we can try to control the formation of this then we will be able to limit the growth of a biofilm. We identified a particular gene WcaF[9] that encodes an acytl transferase which is important in the formation of colanic acid from metabolites.
Formation:
- Begins with attachemnt of free-floating microorganisms at the surface
- Initially there are weak Van Der Waal forces
- If colonists not immediately removed, they can anchor themselves more permanently using cell adhesion molecules
- First colonists facilitate the arrival of other cells by providing more diverse adhesion sites and building matrix that holds the biofilm together
- During colonisation cells communicate through quorum sensing
- Once colonisation has begun biofilm grows through cell-division and recruitment
- Final stage - development: may change in size nad shape
Biofilm formation by E coli on abiotic surface:
- Structures involved in biofilm formation:
- type I pili
- curli for adherence properties - (ompR234 increases the expression of csgA, the curlin-encoding gene porin genes ompC and ompF)
- OmpC porin
- Major role of exopolysaccharides in biofilm development
- Bacteria within biofilms encounter higher-osmolarity conditions, a lower oxygen supply, and higher cell density
- Depending on environmental conditions and biofilm communities, different approaches and cell surface structures such as flagella, pili, S-fimbrial adhesins, autolysins, and curli could be used to initiate biofilm formation
Source:
Abiotic Surface Sensing and Biofilm-Dependent Regulation of Gene Expression in Escherichia coli
Claire Prigent-Combaret, Olivier Vidal,† Corinne Dorel, and Philippe Lejeune
The Escherichia coli OmpR/EnvZ two-component regulatory system, which senses environmental osmolarity, also regulates biofilm formation
OmpR234 protein promotes biofilm formation by binding the csgD promoter region and stimulating its transcription. The csgD gene encodes the transcription regulator CsgD, which in turn activates transcription of the csgBA operon encoding curli, extracellular structures involved in bacterial adhesion. Consistent with the role of the ompR gene as part of an osmolarity-sensing regulatory system, E. coli is inhibited by increasing osmolarity in the growth medium. The ompR234 mutation counteracts adhesion inhibition by high medium osmolarity; ompR234 mutation promotes biofilm formation by strongly increasing the initial adhesion of bacteria to an abiotic surface. Complex regulatory network controls initial adhesion and biofilm formation in Escherichia coli via regulation of the csgD gene.
- Prigent-Combaret C, Brombacher E, Vidal O, Ambert A, Lejeune P, Landini P, and Dorel C. Complex regulatory network controls initial adhesion and biofilm formation in Escherichia coli via regulation of the csgD gene. J Bacteriol. 2001 Dec;183(24):7213-23. DOI:10.1128/JB.183.24.7213-7223.2001 |
Regulation
Indole can act as an extracellular signal to regulate biofilm formation of Escherichia coli and other indole-producing bacteria. Indole may act as a signalling molecule to regulate the expression of adhesion and biofilm-promoting factors
- Martino PD, Fursy R, Bret L, Sundararaju B, and Phillips RS. Indole can act as an extracellular signal to regulate biofilm formation of Escherichia coli and other indole-producing bacteria. Can J Microbiol. 2003 Jul;49(7):443-9. DOI:10.1139/w03-056 |
Abiotic surface sensing and biofilm-dependent regulation of gene expression in Escherichia coli.
- Prigent-Combaret C, Vidal O, Dorel C, and Lejeune P. Abiotic surface sensing and biofilm-dependent regulation of gene expression in Escherichia coli. J Bacteriol. 1999 Oct;181(19):5993-6002. DOI:10.1128/JB.181.19.5993-6002.1999 |
- Di Martino P, Merieau A, Phillips R, Orange N, and Hulen C. Isolation of an Escherichia coil strain mutant unable to form biofilm on polystyrene and to adhere to human pneumocyte cells: involvement of tryptophanase. Can J Microbiol. 2002 Feb;48(2):132-7. DOI:10.1139/w02-001 |
Strains of E coli
- uvrY
- BarA/uvrY
- EnvZ/OmpR
- cpx
Applications:
- Biofilms found on the hull of a ship consist of large organisms like barnacles, mussels, and host of other zooplankton and phytoplankton. These biofilms slow a ship and are expensive to remove and prevent. Current methods to prevent biofilm formation on ships include a wide variety of toxic marine paints. However, these paints tend to wear off and biofilms which are resistant form on them without regard to the toxins.
- barnacles physically corrode the hull and increase drag on the ship - thus just need to be kept away from the ship
- ways to repel barnacles:
- paint that chemically repels barnacles
- zinc oxide
- Mussel adhesive protein (Mgfp-5): form very strong and permanent bonds under water
- need a method to degrade Mgfp-5
- avaiable mussel repellenta are toxic - cause water pollution
- paints that slowly release copper in the water - pollution
- mussels have the hardest time adsorbing onto hydrophobic surfaces (http://www.calpoly.edu/~deastlic/surface_chem/home.html)
- Hwang DS, Yoo HJ, Jun JH, Moon WK, and Cha HJ. Expression of functional recombinant mussel adhesive protein Mgfp-5 in Escherichia coli. Appl Environ Microbiol. 2004 Jun;70(6):3352-9. DOI:10.1128/AEM.70.6.3352-3359.2004 |