IGEM:Hong Kong HKUST/Investigations/Identifying candidate colonies in molecular level/Entry Base
<html> <body>
Identifying Candidate Colonies in Molecular Level
</body> </html>
Authors
- Cheng, Karen K.O.
- Tam, Phoebe L.F.
- Tam, Sabrina K.M.
- Tang, Mandy L.Y.
- Siu, Mona M.Y.
- Yiu, Stephanie P.T.
Abstract
This training is to recap the basic steps and procedures of making constructs, such as digestion, ligation and transformation. In this training,constructs is made by replacing mRFP with GFP in the parental plasmid. Both plasmid was digested with XbaI and SpeI. Hence, in the product, five different possible constructs may resulted: desired constructs, constructs with wrong insert orientation, constructs with two backbones, constructs with two inserts and self-ligated plasmids. The aim of making these constructs is to use methods identifying the candidate colonies with the desired sequence. Colony PCR have been chosen to identify the colonies. To distinguish the orientation, reverse primer and VF2 have been used. While distinguish between self-ligated plasmid and the constructs with two backbone or inserts, VF2 and VR primers are used. After running the PCR products in gel electrophoresis, the one showing the expected band size are concluded having the desired construct.
Colonies are transformed and grown after cloning. However, colony PCR are concluded to be failed as the positive control did not shown band in gel electrophoresis. Hence, colonies with the desired constructs can not be distinguished. Further experiments is needed for identification.
Introduction
Identifying candidate colonies, people usually clone the constructs with reporters, so that colours can be observed from the colonies directly with bare eyes. However, this cannot show whether the orientations is correct or not. It has to be checked in molecular level. In this investigation, insert(GFP) and backbone were digested with the enzymes XbaI and SpeI, thus, the mRFP could be replaced by GFP but there would be five possible results due to different combinations of orders between the insert and backbone. In order to identify the correct one, colony PCR was used followed by gel electrophoresis.
Colony PCR can be used to check whether the constructs contain the desired insert or not. There are three main stages for the colony PCR, including denaturation step, annealing step and extension step. Plasmid DNA can be released to act as the template for amplifying the targeted DNA sequences during the denaturating step (initial heating step) while the oligonucleotide primer hybridize to the single DNA strands during the annealing step. For the extension step, engineered DNA polymerase will add complementary bases to the single DNA strands to build double-stranded DNA. These efficient steps can actually lift the throughput of colony PCR. Aside from this, colony PCR can also be used to determine the insert orientation and check if there is self-ligation by inserting specific primers which can provide the information on the molecular size. Meanwhile, no PCR amplicon exists unless the orientation is correct.
These properties can be made use of in this investigation to identify candidate colonies in molecular level. Two different pairs of primers were applied to produce two sets of results separately for checking the orientation and self ligation after digestion and ligation.
Methods and Materials
To study the methods of identifying candidate colonies in molecular level, a piece of DNA insert containing a GFP generator and another plasmid backbone containg a mRFP gene were restricted by XbaI and SpeI. The DNA fragments were then ligated and transformed. To verify the success of ligation, a colony pcr was performed followed by gel electrophoresis. Then, the results are compared with negative and positive controls to select cells containing the desired DNA sequence.
2 sets of similar experiments were carried out.
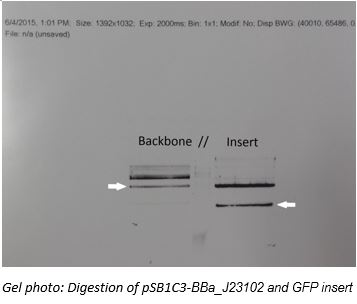
The concentration of pSB1C3-BBa_J23102 and GFP generator were accessed by NanoDrop 2000 spectrophotometer. Approximately 2000ng of BBa_J2310 was digested with 0.2ul of XbaI (NEB, R0145L) and 0.2ul of SpeI-HF (NEB, R0140L) in CutSmart buffer. Similarly, approximately 6000ng of GFP generator was digested with 0.2ul of XbaI (NEB, R0145L) and 0.2ul of SpeI-HF (NEB, R0140L) in CutSmart buffer.Reactions were incubated at 37˚C for 1.5 hour. DNA samples obtained from digestion were analyzed by running 1% agarose gel electrophoresis with 120V electricity for 45 minutes, which was pre-stained by Midori Green for visualization of DNA fragments. Insets were extracted out from the gel and purified using Favorgen FavorPrep™ GEL/PCR Purification Mini Kit.
The digested BBa_J23102 and GFP generator were then ligated with 0.5ul T4 DNA ligase, in 10X T4 ligase buffer. Reactions were incubated at room temperature for 1 hour and underwent transformation, followed by overnight incubation.
26 colonies were picked for colony PCR. The colonies were first mixed with 0.85% NaCl for bursting the colonies.Three sets of control setups were included, which are competent cell(C), plasmid with mRFP(M) and plasmid with GFP(G). Each 2 μl of colony soultion was mixed with 9 μl of water, 4 μl of 1mM dNTP, 4 μl of buffer, 0.01 μl of 75X Taq Pol, 0.5 μl of forward primer and 0.5 μl of reverse primer. For Set 1, VF2 and VR were the forward and reverse primers. For Set 2, VF2 and reverse primer of the insert were the forward and reverse primers.
Both sets underwent colony PCR, with 8 minutes of 95°C initial denaturing, followed by 45 seconds of 95°C denaturation, then 45 seconds of 55°C annealing, and 3 minutes and 20 seconds of 68°C extension. A total of 20 cycles were repeated. A 10-minute 72°C final extension was carried out and holding at 10°C.
A gel elctrophoresis was then carried out to select the desired fragments by comparing the experimental setups with control setups. 1% gel was used for Set 1 and 1.5% gel was used for Set 2, , which were both pre-stained by Midori Green for visualization of DNA fragments. The two gels underwent electrophoresis with 140V electricity for 40minutes.
Rationale
XbaI and SpeI restriction enzymes were being used for digestion, therefore, sticky ends of XbaI and SpeI were made. As XbaI and SpeI has a similar restrction sequence, it is likely for them to have ligations, which may in turn results in self-ligation and reverse orientation . In order to identify the candidate colonies, colony PCR was performed, and gel electrophoreiss was then adopted for identifying the candidate colonies.
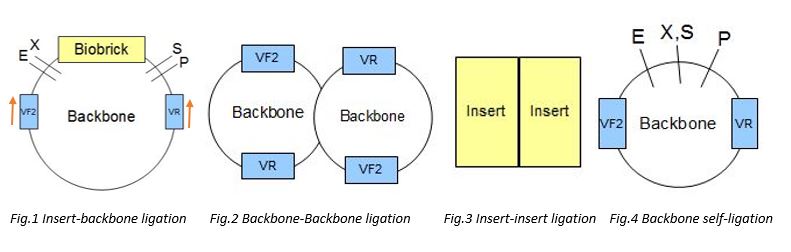
For checking self ligation Primers of VR and VF2 were being used. The two primers would bind to the corresponding VR and VF2 binding sites on the backone, and amplify all the seqeunces in between the VR and VF2 sites. (Fig.1) There would be 4 possible outcomes, including backbone-backbone ligation (Fig.2), insert-insert liagtion (Fig.3), backbone self-ligation (Fig.4) and the insert-backbone ligation (Fig.1). First, for backbone-backbone ligation, as there are only 2 backbones ligate with each other without the insert, the VR and VF2 primers would only amplify the sequences of the biobrick prefix and suffx, as well as a few sequence in between the VR and VF2 sites, therefore, after gel elcectrophoresis, the expected band size would be around 300-400bp. Second, for insert-insert ligation, as the two inserts ligate with each other without the backbone, there would be no VR and VF2 sites and so, the primers would not bind to it and wouldnot have amplification. Therefore, after the gel electrophoresis, the expected band size would be 0bp. Third, for the backbone self ligation, as the backbone ligate with itself without the insert, the VR and VF2 primers would only amplify the sequences of the biobrick prefix and suffx, as well as a few sequence in between the VR and VF2 sites, therefore, after gel elcectrophoresis, the expected band size would be around 100-200bp. Forth, for backbone-insert ligation, after the gel electrophoresis, the expected band size would be around 2000bp.
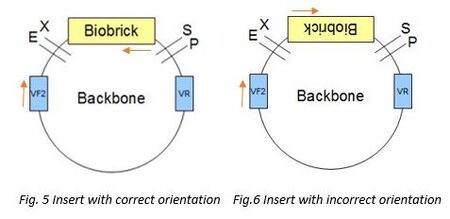
For checking orientation VF2 and the reverse primer of GFP were being used. VF2 primers would bind to the VF2 site on the backbone while the reverse primer will bind to the antisense of the GFP insert. The 2 primers would then amplify all the sequence between the insert and VF2 sites. There would be two possible outcomes, one with insert that is of correct orientation (Fig.5), another with insert of reverse orientation (Fig.6). First, for the product with insert of correct orientation, after gel electrophoresis, the expected band size would be around 1300bp. Second, for the product with insert of reverse orientation, as the two primers would not meet each other, as a result, there would be no amplification, therefore, after the gel electrophoresis, the expected band size would 0bp.
Results and Interpretations
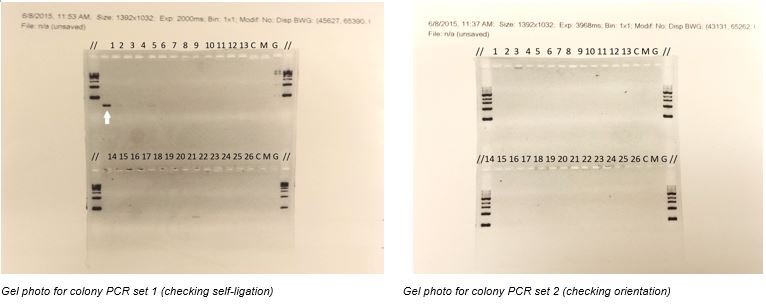
In the set 1 colony PCR gel photo, there is a band of size around 300-400bp resulted, it is suspected to be the backbone-backbone ligation outcome. In the set 2 colony PCR gel photo, no observable bands can be found.
As there are no bands for the positive control in both the gel photos, the PCR reaction could not be proved to have worked. As a result, the colony PCR is considered to be failed.
Discussion
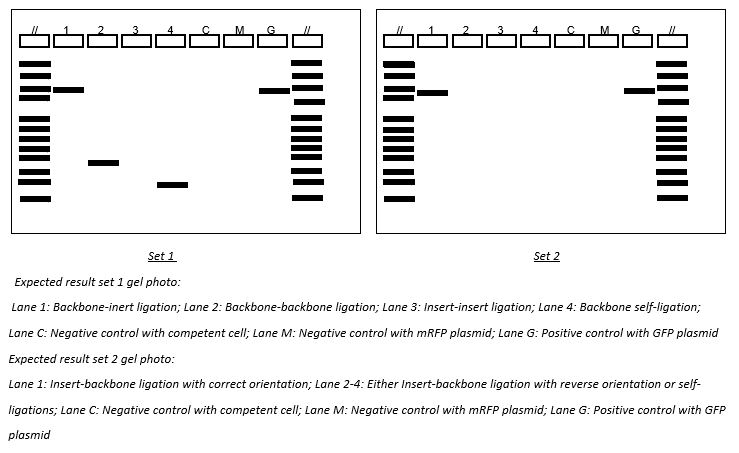
As from the gel photos, positive control of both sets did not show a band. Set 2 even did not show any bands. Hence, it is concluded that the colony PCR experiment was failed. The illustration below is the expected results if colony PCR was successful:
From the illustration of set 1, Lane 1 shows a band of size around 2000bp, which represents an outcome of backbone-insert ligation with unknown orientation. For Lane 2, it shows a band of size around 300-400bp, which represents an outcome of backbone-backbone ligation. For Lane 3, it does not show a band, which represents an outcome of insert-insert ligation. For Lane 4, it shows a band of size around 100bp, which represents an outcome of backbone self-ligation.
From the illustration of set 2, Lane 1 shows a band of size around 1300bp, which represents an outcome of backbone-insert ligation with correct orientation. For Lane 2 to 4, they do not show any bands, which represent outcomes of either backbone-insert ligation with reverse orientation or outcomes with self-ligation.
Improvements
There are few reasons that may lead to the failure of colony PCR. Points below are some ways to improve the experiment. First, pay more attention with the PCR mixture to ensure the mixture are included with all necessary materials and the right amount. Secondly, add more colony solution or plasmid to the PCR mixture so that the initial DNA concentration is higher to bring out a more concentrated PCR product.
Another method can be use to identify candidate colonies is by restriction check. Using two enzyme which cuts the insert and the backbone respectively, will be able to check the orientation with corresponding restricted bang size.
Conclusion
Cloning have been carried out to construct a plasmid by replacing mRFP with GFP. However, as both sets of PCR products fail to show bands in our Positive control, it is concluded that this is a fail PRC experiment. Hence,and due to time constrains, colonies containing which type of possible constructs can not be distinguished. Further experiments are needed for identification of candidate colonies.