IGEM:Harvard/2008/Lab Notebooks/DailyBook/Week1/Chemical and Light
Chemical
Results from Friday (6/20) Electroporation and Transformation in S1
6/23:
Name Registry Name Description Origin Marker Observations P1a J23113 Low promoter GFP tester pMB1 Kan 16 pink colonies of approx 7mm diameter P2a J23150 Medium promoter GFP tester pMB1 Kan no growth P3a J23151 High promoter GFP tester pMB1 Kan 8 pink colonies of approx 7mm diameter P12 pETDuet-1 Low copy plasmid pBR322-derived ColE1 Amp non-specific Lawn; restreaks did not grow P13 pCDFDuet-1 Low copy plasmid CloDF13 Sm 41 pink colonies and some small yellow ones that may be E. coli P14 pCOLADuet-1 Low copy plasmid ColA Kan No growth
Growing cultures of E. coli and S1 with BioBrick Parts or Duet Vectors in them
6/23:
Picked colonies of E. coli and S1 and grew overnight at 37 and 30 degrees, respectively.
Strain Plasmid Name Registry Name Plate Information Marker Colonies Picked E. coli P15 A1 6/18 electrocompetent Cm 2 TOP10 P4 pACYC duet 6/19 restreak electrocomp Cm 2 TOP10 P7 pSB1A2 6/19 restreak electrocomp big and small Cm 2 (1 from big, 1 from small) TOP10 P8 BBa_J04450 6/19 restreak electrocomp big and small Amp 2 (1 from big, 1 from small) TOP10 P9 BBa_J04430 6/19 restreak electrocomp big and small Amp 2 (1 from big, 1 from small) TOP10 P10 BBa_I715038 6/19 restreak electrocomp big and small Amp 2 (1 from big, 1 from small) S1 P4 pACYC duet 6/18 electrocomp Cm 2 S1 P21 A1 6/19 electrocomp Cm 2 S1 P13 pCDFDuet-1 6/20 electrocomp Sm 2 S1 P1a J23113 6/20 electrocomp Kan 3 S1 P3a J23151 6/20 electrocomp Kan 3
Total number of cultures: 24
6/24:
None of the samples in the LB amp media grew. All the others grew and then were miniprepped and stored in glycerol.
- Glycerol stocks: 250 uL of culture in 500 uL of 20% LB glycerol.
Concentrations of DNA from minipreps:
Plasmid Organism Concentration (ng/uL) P1A a S1 92.54 P1A b S1 55.34 P1A c S1 58.21 P3A a S1 62.43 P3A b S1 168.41 P3A c S1 151.49 P4 S1 150.32 P13 S1 435.39 P21 S1 219.33 P4A a TOP10 49.81 P4A b TOP10 47.22 P21A a TOP10 61.16 P21A b TOP10 55.71
6/26: Amp cultures still did not grow in liquid media, neither at 1:1000 or a 1:2000 dilution of amp
Transformation of P5 and P6 into E3
6/24:
P5 and P6 were punched from the book and transformed in E3 (the death resistant strain)
6/25: No Growth
16S PCR to Identify S1 and E. Coli
6/24:
Performed PCR as per protocol listed in General Protocols.
Primer Sets
- 1 = Shewanella F and Shewanella R
- 2 = E. coli F and E. coli R
- 3 = Common F and Common R
Tube # Strain/ Colony of Template DNA Primer Set Used Volume of Template DNA Volume of Primer Volume of Supermix Volume of Water 1 S1 P1a 1 (SF & SR) 1uL 1uL (each) 45uL 2uL 2 S1 P1a 1 (SF & SR) 3uL 1uL (each) 45uL 2uL 3 S1 P1a 2 (EF & ER) 1uL 1uL (each) 45uL 2uL 4 S1 P1a 2 (EF & ER) 3uL 1uL (each) 45uL 2uL 5 S1 P1a 3 (CF & CR) 1uL 1uL (each) 45uL 2uL 6 S1 P1a 3 (CF & CR) 3uL 1uL (each) 45uL 2uL 7 S1 P3a 1 (SF & SR) 1uL 1uL (each) 45uL 2uL 8 S1 P3a 1 (SF & SR) 3uL 1uL (each) 45uL 2uL 9 S1 P3a 2 (EF & ER) 1uL 1uL (each) 45uL 2uL 10 S1 P3a 2 (EF & ER) 3uL 1uL (each) 45uL 2uL 11 S1 P3a 3 (CF & CR) 1uL 1uL (each) 45uL 2uL 12 S1 P3a 3 (CF & CR) 3uL 1uL (each) 45uL 2uL 13 S1 MR-1 1 (SF & SR) 1uL 1uL (each) 45uL 2uL 14 S1 MR-1 1 (SF & SR) 3uL 1uL (each) 45uL 2uL 15 S1 MR-1 2 (EF & ER) 1uL 1uL (each) 45uL 2uL 16 S1 MR-1 2 (EF & ER) 3uL 1uL (each) 45uL 2uL 17 S1 MR-1 3 (CF & CR) 1uL 1uL (each) 45uL 2uL 18 S1 MR-1 3 (CF & CR) 3uL 1uL (each) 45uL 2uL 19 E. coli P1a 1 (SF & SR) 1uL 1uL (each) 45uL 2uL 20 E. coli P1a 1 (SF & SR) 3uL 1uL (each) 45uL 2uL 21 E. coli P1a 2 (EF & ER) 1uL 1uL (each) 45uL 2uL 22 E. coli P1a 2 (EF & ER) 3uL 1uL (each) 45uL 2uL 23 E. coli P1a 3 (CF & CR) 1uL 1uL (each) 45uL 2uL 24 E. coli P1a 3 (CF & CR) 3uL 1uL (each) 45uL 2uL
6/25
Gel results from PCR:
Key:
Well # Strain/ Colony Primer Set Used Volume of Template DNA 1 1 Kb ladder N/A N/A 2 S1 P3a 1 (SF & SR) 3uL 3 S1 P3a 1 (SF & SR) 1uL 4 S1 P3a 2* (EF & ER) 1uL 5 S1 P3a 3 (CF & CR) 1uL 6 S1 P3a 3 (CF & CR) 3uL 7 S1 P1a 1 (SF & SR) 3uL 8 S1 P1a 1 (SF & SR) 1uL 9 S1 P1a 2 (EF & ER) 1uL 10 S1 P1a 2 (EF & ER) 3uL 11 S1 P1a 3 (CF & CR) 3uL 12 S1 P1a 3 (CF & CR) 1uL
Note: S1 P3a With Primer Set 2 and 3 uL of template DNA evaporated during PCR so was not included in the gel.
Key:
Well # Strain/ Colony Primer Set Used Volume of Template DNA 1 1 Kb ladder N/A N/A 2 S1 MR-1 1 (SF & SR) 1uL 3 S1 MR-1 2 (EF & ER) 1uL 4 S1 MR-1 3 (CF & CR) 1uL 5 100 bp Ladder N/A N/A 6 E.coli P1a 1 (SF & SR) 1uL 7 E.coli P1a 1 (SF & SR) 3uL 8 E.coli P1a 2 (EF & ER) 1uL 9 E.coli P1a 2 (EF & ER) 3uL 10* E.coli P1a 3 (CF & CR) 1uL 11 E.coli P1a 3 (CF & CR) 3uL 12 1 Kb Ladder N/A N/A
Note: There was a problem with the gel in Well 10, which most likely accounts for the lighter band.
Remy's pZ vector system Plasmids
6/25: transfer colonies to liquid media
6/26: mini prep plasmids
Name Registry Name Description Origin Marker Regulatory unit Concentration ng/uL P27 pZS4int2 repressor plasmid for chromosomal integration pSC101 Sm Constitutive expression of LacI and TetR P28 pZE21 GFP ColE1 Kan PLtctO-1 P29 pZS*2R Venus (mutant YFP) pSC101* Kan PL (Plambda) P30 pZA32 YFP p15A Cm PLlacO-1 P31 pZS*1R Venus pSC101* amp PL (Plambda) P32 pZA32 Venus p15A Cm PLlacO-1 P33 pZA31 Venus p15A Cm PLtctO-1 P34 pZE12 GFP ColE1 Amp PLlacO-1 P35 pLDR8 Accessory for chromosomal integration ?? ??? ??
Also two strains with integrated copies of repressors, low level constitutive promoters
- MC4100 Z1 - repressor lac and tet (Sm resistant)
- DH10B Z2 - repressor lac and tet (Sm resistant)
Re-Transformation of BioBrick Parts in E1 and E3
6/26:
Re-transformed BioBrick parts in E1 because amp plates weren't working. Also re-transformed P5 and P6 in E3.
Name Registry Name Description Origin Marker Transformed? (6/26) Picked colonies? Miniprepped? Made glycerol stocks? P7 pSB1A3* High copy plasmid with Amp resistance ColE1 Amp Failed in E1 P8 BBa_J04450 RFP with LacI promoter pMB1 Amp Failed in E1 P9 BBa_J04430 GFP with LacI promoter pMB1 Amp Failed in E1 P10 BBa_I715038 T7 Polymerase with LacI promoter pMB1 Kan/Amp Failed in E1 P36 BBa_I763004 GFP + IPTG + LVA Kan/Amp Failed in E1 P5 (2007) pSB3K3* Low-Medium copy vector (w/ death gene) p15A Kan Yes, in E3 2 (A and B spilled) P6 (2007) BBa-E1010 RFP only (w/ death gene) pMB1 Kan Failed in E3 P5 (2008) pSB3K3* Low-Medium copy vector (w/ death gene) p15A Kan Failed in E3
Notes:
- Control was also done with untransformed E3 on LB plates.
- pSB1A3 was used instead of pSB1A2 because the Parts Registry lists pSB1A3 as the replacement for pSB1A2.
- P5 and P6 were taken from Spring 2007 plates, and iGEM 2008 P5 was punched out and transformed again.
6/27:
P5 (2007) plate was put in incubator for longer, and will be picked on Sunday along with the other transformations done today.
Name Registry Name Description Origin Marker Transformed? (6/27) Picked colonies? (6/29) Miniprepped? Made glycerol stocks? P12 pETDuet-1 Low copy plasmid pBR322-derived ColE1 Amp in E1 2 P37 BBa_I722007 Constructive expression LacI w/ pTetR promoter Amp in E1 2 (A and B didn't work) P38 BBa_J23114 High constitutive promoter Amp in E1 2 P38 (2007) BBa_J23114 High constitutive promoter Amp in E1 2 (A and B didn't work) P39 BBa_J23113 Low constitutive promoter Amp in E1 2 (B didn't work) P39 (2007) BBa_J23113 Low constitutive promoter Amp in E1 2 (B did not work) P40 BBa_B0032 Ribosome Binding Site Amp in E1 2 (B didn't work) P40 (2007) BBa_B0032 Ribosome Binding Site Amp in E1 2 (A did not work) P41 BBa_B0010 Transcriptional Terminator Amp in E1 2 (A and B didn't work) P41 (2007) BBa_B0010 Transcriptional Terminator Amp in E1 2 (Neither worked) P42 BBa_C0012 LacI coding region Amp in E1 2 (A and B didn't work) P42 (2007) BBa_C0012 LacI coding region Amp in E1 1 (didn't work) P43 [1] TetR coding region Amp in E1 2 (A and B didn't work) P43 (2007) [2] TetR coding region Amp in E1 2 (B didn't work) P44 BBa_C0051 cI lambda Amp in E1 2 (A and B didn't work) P44 (2007) BBa_C0051 cI lambda Amp in E1 2 (B didn't work) P45 BBa_E0240 GFP only w/ RBS & terminator Amp in E1 2 (A and B didn't work) P45 (2007) BBa_E0240 GFP only w/ RBS & terminator Amp in E1 2 P46 BBa_I51020 Base Vector Amp in E3 2 P47 BBa_P1003 Kan Resistance Cassette Kan No in E1 0 P47 (2007) BBa_P1003 Kan Resistance Cassette Kan in E1 2 (A and B spilled) P48 BBa_P1004 Cm Resistance Cassette Cm No in E1 0 P48 (2007) BBa_P1004 Cm Resistance Cassette Cm in E1 2 P49 BBa_P1002 Amp Resistance Cassette Amp in E1 2 (B didn't work) P49 (2007) BBa_P1002 Amp Resistance Cassette Amp in E1 2 P50 BBa_P1005 Tet Resistance Cassette Amp in E1 2 (A and B didn't work) P50 (2007) BBa_P1005 Tet Resistance Cassette Amp in E1 2
McConkey Plates to Distinguish E. coli from Shewanella
6/24
Plates streaked and put in incubator.
6/25:
Top Left: E1
Bottom Left: E1-P1a
Top Right: S1
Bottom Right: S1-P1a
Top Left: chemically competent E. coli
Bottom Left: E1-P3
Top Right: WT S1
Bottom Right: S1-P3
Top Left: E1
Bottom Left: E1-A1 (P21)
Top Right: S1
Bottom Right: S1-A1 (P21)
Growing Shewie anaerobically
6/25:
Added lactate and birnesite to LB in a vial, bubbled N2 gas to degas it and then added Shewie at concentrations of 100 uL and 500 uL, while keeping a control with no Shewie added. Vials were placed in 30 degree shaking incubator overnight.
6/26:
All vials, including the control, turned light yellow (the color of the LB). This was most likely due to contamination, which was partly confirmed by looking at the vials through the microscope. This was not unexpected because the vials we used had not been autoclaved, and it was very difficult to maintain sterile conditions and precise concentrations of birnesite/ lactate.
Questions/ Concerns
Regarding transformations of BioBrick Parts:
- Given the parts that we've succeeded in transforming in either E. coli, or Shewanella, or both, what is our plan for designing vectors? How many plasmids will we need?
- How should we incorporate Remy's plasmids into those designs?
- To what extent can we take parts from or use the Duet vectors?
- Can we confirm that pMB1 does work in Shewanella, contrary to the 1997 paper that suggests otherwise?
- If we have successful transformations with both the 2007 and 2008 versions of a BioBrick part, should we prefer one over the other?
Regarding anaerobic Shewie growth: (Many of these questions may be the same as those posed by the Widgetry group.)
- What are the optimal concentrations of lactate and birnesite that we should use?
- Tips on maintaining sterility/ preventing contamination?
- Orianna's paper lists three different media that she used for various purposes-- what does our basic media need?
- How much Shewie should we be using? Should we be adding them at a certain growth phase or OD?
Light
Transformations of P11, P15-20
ENDED IN FAILURE: SEE NEW TRANSFORMATION BELOW
Colony counts
6/23:
| Name | Description | Origin | Size | Marker | Number of Colonies |
| P11 | Light responsive system, dual regulation | pUC19-derived pMB1 | 4333 | Amp/Cm | 224 |
| P15 | GFP with Tet promoter | ColE1 | 937 | Amp | 360 |
| P16 | Tet repressible promoter | pMB1 | 54 | Amp | 264 |
| P17 | Inverter (TetR w/o promoter and Tet promoter) | OriS, P1 lytic, F1 | 902 | Kan | 2 |
| P18 | lambda promoter (cI regulated) | pMB1 | 49 | Amp | 112 |
| P19 | RFP with lambda promoter | pMB1 | 918 | Amp | 104 |
| P20 | GFP (LVA tagged) with Lac promoter | pMB1 | 1122 | Amp, Kan | 136 |
Restreaking
06/23: P15, which has GFP under a constitutive Tet promoter did not appear to fluoresce (none of the fluorescent constructs did), so we restreaked all the of the plates (with the same antibiotic on which they were originally grown). They were left in the 37 degree incubator along with a blank Kan and a blank Amp plate (- controls).
06/24: All of the restreaked plates grew and were streaked to individual colonies. Both the Kan and Amp negative controls (no cells) were blank. However, p15 (GFP under pTet) did not appear to glow.
Overnight cultures
06/23: The above was repeated in 5mL LB liquid cultures instead of plates.
- 06/24: None of the cultures in LB Amp grew (E1P17 in LB Kan did grow). We recultured the other samples in LB Amp and made a master plate with patches from the same colonies that were cultured. Since others seemed to get similar problems (no growth in liquid culture but growth on plates for Amp), we made fresh LB Amp (from freshly made Amp stock) and repeated the cultures.
- 06/25: These LB-Amp cultures did not grow in the new medium either. We plated them on new LB amp plates (new plates 1X amp or 1/1000) and cultured them in low concentration LB amp (0.5X or 1/2000 amp).
- 06/26: None of the cultures grew on either of the liquid media (1X and .5X LB-Amp). Additionally, none of the master plate colonies restreaked onto the new LB-Amp plates that we poured grew. This suggests to us that the original LB-Amp plates were not selective enough (perhaps Amp broke down). As a control that our bacteria were capable of growth, we inoculated a plain LB liquid culture with each of the 6 colonies from the master plate. This culture did grow (cloudy). A plain LB liquid culture left as a negative control did not.
Miniprep of P17
ENDED IN FAILURE: SEE NEW TRANSFORMATION BELOW
06/24: Miniprep of E1P17. The DNA is in the -20 °C iGEM freezer. The DNA concentration is 54.8 ng/μL.
Restriction Digest
06/24: Digest of P17 DNA with Xba1 and Spe1. The entire plasmid is 4425 bp and the short fragment (containing only the BioBrick) is 902 bp.
Digest mixture (incubated overnight at 37 °C):
15 μl DNA
6.25 μl water
2.5 μl NEB Buffer 2
0.25 μl 100X BSA
1 μl Xba1
1 μl Spe1
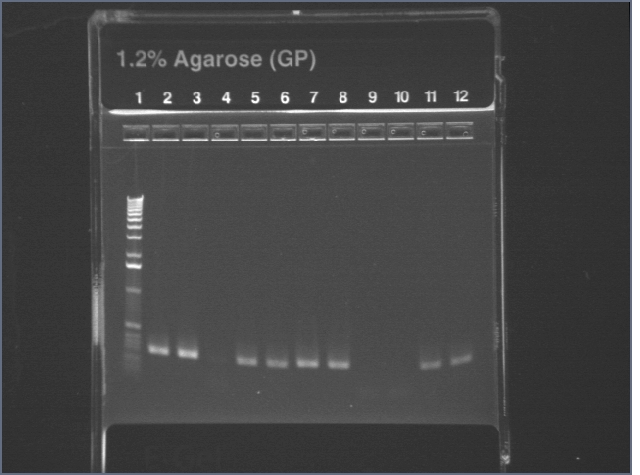
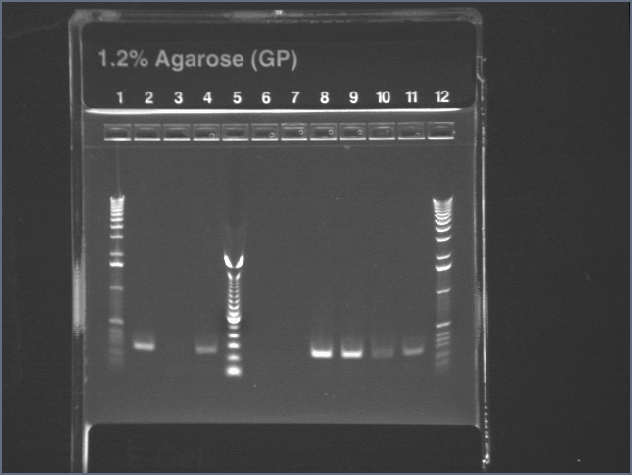
Gel of digest products (6/25)
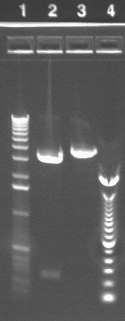
|
1.2% agarose E-gel run for 30 min and visualized using EtBr/UV | |
|---|---|---|
| Lane | Contents | |
| 1 | 1 KB ladder | |
| 2 | Digest. Note that these are not what would be expected for the correct plasmid (902 band for part and 4.5kb for vector). Even if there are cutting issues, the sum of the band sizes is around 2kb too low. | |
| 3 | Undigested plasmid | |
| 4 | 100 bp ladder | |
06/25: Since the previous digest was unsuccessful, we digested the same plasmid with different combinations of restriction enzymes. We also tested two other plasmids from the chemical group (P4 and P21). The digest mixtures were the same as above (6/24) with the appropriate enzyme swapped in. For the single digests, the volume of water was 6.75 μl and all else remained constant. Again, incubation is overnight at 37 °C.
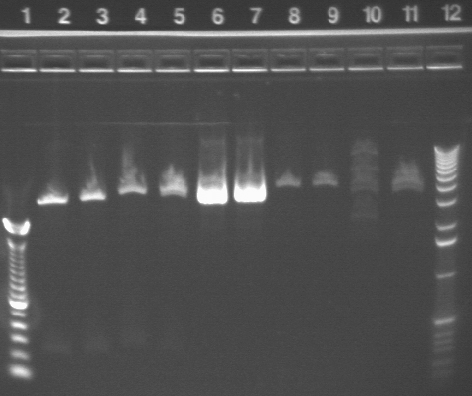
|
1.2% agarose E-gel run for 30 min and visualized using EtBr/UV | |
|---|---|---|
| Lane | Contents | |
| 1 | 100 bp ladder | |
| 2 | P17 with XbaI, SpeI | |
| 3 | P17 with EcoRI, SpeI | |
| 4 | P17 with EcoRI, PstI | |
| 5 | P17 with XbaI, PstI | |
| 6 | P17 with XbaI | |
| 7 | P17 with SpeI | |
| 8 | P4 with XbaI, SpeI | |
| 9 | P4 with EcoRI, SpeI | |
| 10 | P21 with XbaI, PstI | |
| 11 | P21 with EcoRI, PstI | |
| 12 | 1 KB ladder | |
From the lanes with P17, which again indicate that the plasmid we have from the miniprep is not the right size, we conclude that the P17 transformation failed. It is being repeated (see below).
Transformations of P22-24
ENDED IN FAILURE: SEE NEW TRANSFORMATION BELOW
06/24: Transformations occurred in duplicate (TOP10 and DH5α) with Amp selection.
06/25: All plates had colonies. We inoculated liquid cultures of cells with each of these plasmids in low (0.5X or 1/2000 amp) and high (1X or 1/1000 amp) concentration LB amp.
E1P22: 376 colonies E1P23: 352 colonies E1P24: 392 colonies E2P22, E2P23, and E2P24 had too many colonies to count.
- 06/26: None of the liquid cultures (both high and low Amp) had any growth. Additionally, we patched the colonies we were growing on a master plate that was of the new LB-Amp plates that we poured. No colonies grew on this plate either. This again seems to indicate that the initial transformation plates did not have enough selection (perhaps Amp was degraded).
Transformations of P17, 20, 25, 26
SEE NEW TRANSFORMATION BELOW
06/25: Transformed P17, P20, P25, and P26 in DH5α cells. P17 and P20 have Amp and Kan resistance, but since AmpR cells don't seem to grow in the liquid media, we're repeating the process by plating the cells on Kan (and eventually grow them on LB-Kan liquid medium). P25 and P26 are new and have Amp resistance.
06/26: P20 (kan plate) had 4 colonies and P25 (amp plate) had 1 colony. The other plates (with P17, P20, and P26) did not have any colonies. The kan and amp negative control plates (DH5α cells with no DNA) also had no colonies.
New transformations: P11, P15-20, P22-26
06/26
We transformed TOP10 cells using parts from both the 2008 and 2007 registries, apart from P11 and P19 which were not available in the 2007 registry.
The protocol for getting DNA from the 2007 registry is as follows:
1. Puncture a hole in the foil with a pipet.
2. Add 15 μl H2O and wait for about 5 minutes.
3. Add 1 μl DNA to 50 μl bacteria.
P17 and P20 were plated on Kan plates. All other samples were on Amp plates that we made (at 1X or 1/1000 amp).
Transformation results
Both negative plate controls (Amp and Kan) did not have any colonies. The "mock transformations" (no DNA) were also negative. The positive plate control had a sample of pEZ12 GFP from Remy and this plasmid also carried Amp resistance. There was a lawn of bacteria on this plate. We also did a positive transformation control that had 1 μg pUC19 DNA for 50 μl TOP10 cells. This DNA came with the TOP10 cells from Invitrogen and there were 26 colonies on the plate.
| Plasmid | Number of Colonies |
| P15 (2007) | 112 |
| P16 (2008) | 2 |
| P18 (2007) | 22 |
| P20 (2007) | 96 |
| P23 (2007) | 40 |
| P24 (2007) | 4 |
| P26 (2007) | 14 |
6/10 of the 2007 parts appear to have transformed (colonies grew) and 1/12 of the 2008 parts appear to have transformed successfully.
Remy's positive control with GFP was fluorescent, along with P15 (GFP with Tet promoter) and P20 (LVA-tagged GFP with Lac promoter). This indicates that the transformations of P15 and P20 were successful.
Set up liquid cultures (with master plates) for all of the plasmids that had colonies.
Minipreps and digests
6/28: The overnight cultures grew successfully and were mini-prepped. Note that there are replicates (e.g. 15A and 15B are different tubes from the overnight cultures. * and *-less denotes replicates during minipreppring). Unfortunately, I forgot to keep culture for glycerol stocks. I performed a NotI digest (see recipe under "RE Digest")- it's the standard single digest recipe, but halfed. The digest ran for the evening, and the results are below:
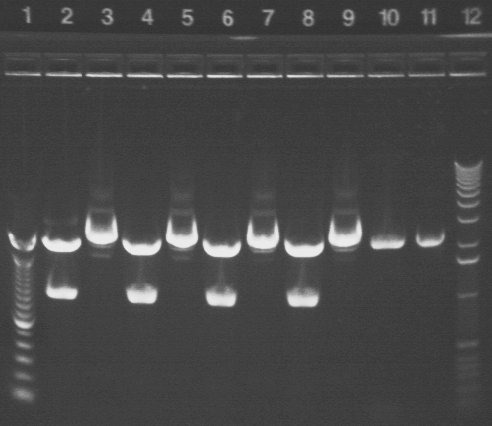
|
1.2% agarose E-gel run for 30 min and visualized using EtBr/UV | |
|---|---|---|
| Lane | Contents | |
| 1 | 100 bp ladder | |
| 2 | P15A* digested with NotI (2079, 937 bp expected) | |
| 3 | P15A* uncut (3016 bp expected) | |
| 4 | P15A digested with NotI (2079, 937 bp expected) | |
| 5 | P15A uncut (3016 bp expected) | |
| 6 | P15B* digested with NotI (2079, 937 bp expected) | |
| 7 | P15B* uncut (3016 bp expected) | |
| 8 | P15B digested with NotI (2079, 937 bp expected) | |
| 9 | P15B uncut (3016 bp expected) | |
| 10 | P16* digested with NotI (2079, 54 bp expected) | |
| 11 | P16* uncut (2133 bp expected) | |
| 12 | 1 KB ladder | |
All of the P15 samples appear to have worked properly. P16 is questionable.
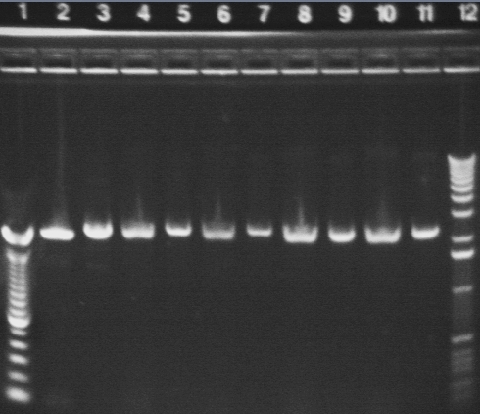
|
1.2% agarose E-gel run for 30 min and visualized using EtBr/UV | |
|---|---|---|
| Lane | Contents | |
| 1 | 100 bp ladder | |
| 2 | P16 digested with NotI (2079, 54 bp expected) | |
| 3 | P16 uncut (2133 bp expected) | |
| 4 | P18A* digested with NotI (2079, 49 bp expected) | |
| 5 | P18A* uncut (2128 bp expected) | |
| 6 | P18A digested with NotI (2079, 49 bp expected) | |
| 7 | P18A uncut (2128 bp expected) | |
| 8 | P18B* digested with NotI (2079, 49 bp expected) | |
| 9 | P18B* uncut (2128 bp expected) | |
| 10 | P18B digested with NotI (2079, 49 bp expected) | |
| 11 | P18B uncut (2128 bp expected) | |
| 12 | 1 KB ladder | |
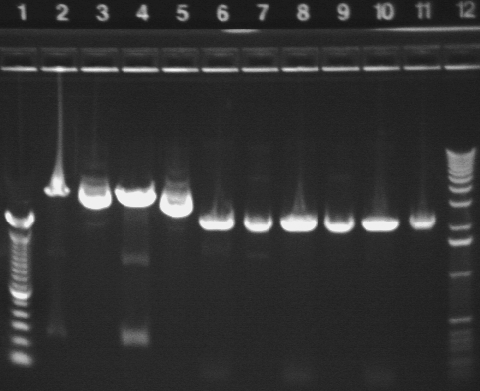
|
1.2% agarose E-gel run for 30 min and visualized using EtBr/UV | |
|---|---|---|
| Lane | Contents | |
| 1 | 100 bp ladder | |
| 2 | P20A* digested with NotI (1122, 3189 bp expected) | |
| 3 | P20A* uncut (4311 bp expected) | |
| 4 | P20A digested with NotI (1122, 3189 bp expected) | |
| 5 | P20A uncut (4311 bp expected) | |
| 6 | P23* digested with NotI | |
| 7 | P23* uncut | |
| 8 | P23 digested with NotI | |
| 9 | P23 uncut | |
| 10 | P23B* digested with NotI | |
| 11 | P23B* uncut | |
| 12 | 1 KB ladder | |
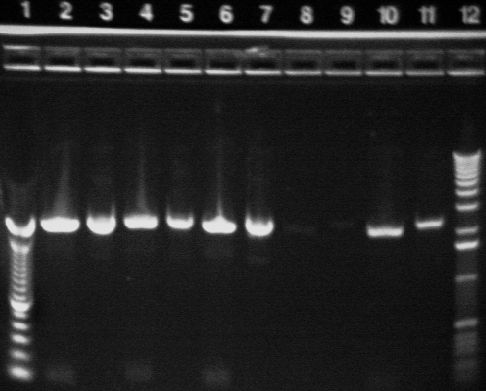
|
1.2% agarose E-gel run for 30 min and visualized using EtBr/UV | |
|---|---|---|
| Lane | Contents | |
| 1 | 100 bp ladder | |
| 2 | P23B digested with NotI | |
| 3 | P23B uncut | |
| 4 | P24A* digested with NotI | |
| 5 | P24A* uncut | |
| 6 | P24A digested with NotI | |
| 7 | P24A uncut | |
| 8 | P24B* digested with NotI | |
| 9 | P24B* uncut | |
| 10 | P24B digested with NotI | |
| 11 | P24B uncut | |
| 12 | 1 KB ladder | |
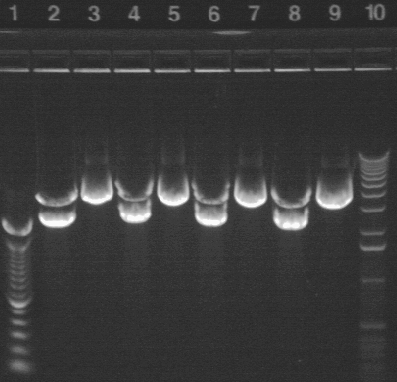
|
1.2% agarose E-gel run for 30 min and visualized using EtBr/UV | |
|---|---|---|
| Lane | Contents | |
| 1 | 100 bp ladder | |
| 2 | P26A* digested with NotI | |
| 3 | P26A* uncut | |
| 4 | P26A digested with NotI | |
| 5 | P26A uncut | |
| 6 | P26B* digested with NotI | |
| 7 | P26B* uncut | |
| 8 | P26B digested with NotI | |
| 9 | P26B uncut | |
| 10 | 1 KB ladder | |
RE digest
06/27
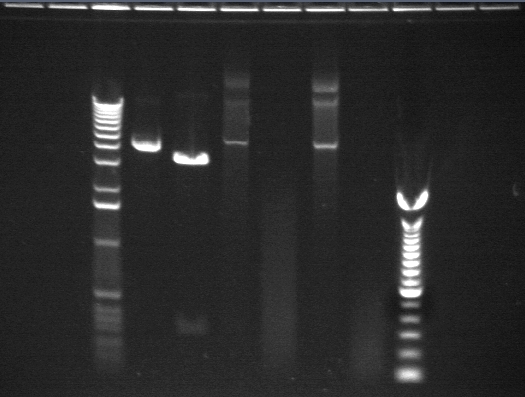
|
1.2% agarose E-gel run for 30 min and visualized using EtBr/UV |
|---|---|
| Lane 3: 1 kb ladder | |
| Lane 4: P17 uncut (should be 4425 bp) | |
| Lane 5: P17 cut with NotI (should be 3503 and 902 bp) | |
| Lane 6: P3 uncut (should be 2750 bp)??? | |
| Lane 7: P3 cut with NotI (should be 1831 and 919 bp)??? | |
| Lane 8: P1 uncut (should be 3002 bp)??? | |
| Lane 9: P1 cut with NotI (should be 2083)??? | |
| Lane 10: 100 bp ladder |
These digests were meant to test the enzymes and digest conditions. P3 and P1 were supposed to be controls. However they were Biobricks miniprepped from Shewie and it doesn't look like the transformation into Shewie worked as even the uncut DNA is not the right size. We will redo the digest with DNA from E. coli.
I set up another overnight digest but doubled the overall volume so that there was 1 μl of the restriction enzyme (NotI).
30 μl DNA
13.5 μl H2O
5 μl 10X NEB buffer 3
0.5 μl 100X BSA
1 μl NotI
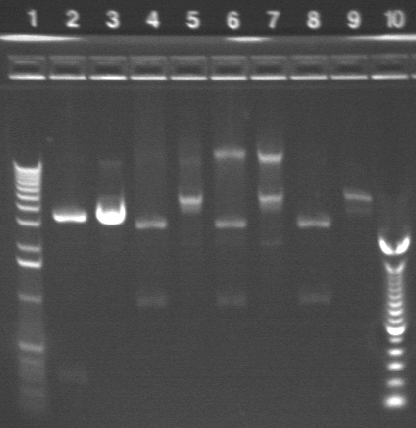
|
1.2% agarose E-gel run for 30 min and visualized using EtBr/UV |
|---|---|
| Lane 1: 1 kb ladder | |
| Lane 2: P17 cut with NotI (should be 3503 and 902 bp) | |
| Lane 3: P17 uncut (should be 4425 bp) | |
| Lane 4: P1 cut with NotI (2750, 919 bp expected) | |
| Lane 5: P1 uncut (3669 bp expected) | |
| Lane 6: P2 cut with NotI (2750, 919 bp expected) | |
| Lane 7: P2 uncut (3669 bp expected) | |
| Lane 8: P3 cut with NotI (2750, 919 bp expected) | |
| Lane 9: P3 uncut (3669 bp expected) | |
| Lane 10:100 bp ladder |
The bands in the cut P1-3 appear to be of the expected size. However, running a sample of the uncut miniprep gives more than one band (is this b/c an unclean miniprep?) P17 continues to digest as if it is a biobrick. However, the bands continue to be of unexpected sizes.