IGEM:Harvard/2006/Cyanobacteria/Notebook/2006-8-25
Miniprep of Nick's cultures #5 and #8
I miniprepped the 4 cultures Nick inoculated last night, and labeled them 5-1, 5-2, 8-1, and 8-2.
From Nick's email:
All three colonies are KaiB constructs. I have made 4 overnight
cultures with the following labels:
"NES 5" is a putative KaiB insert in a J04500 from a 6:1 Insert:Backbone
reaction using the NEB ligase.
"NES 5" is a second culture started from the same colony.
"NES 8" is a putative KaiB insert in a J04500 from a 3:1 Insert:Backbone
reaction using the Roche ligase.
"NES 8" is from a different (*not* clonal) colony from the same ligation
reaction as the other NES 8.
Diagnostic digests of Nick's cultures #5 and #8
| Digest # | Plasmid | Enzymes |
| 1 | 5-1 | X |
| 2 | 5-1 | X, P |
| 3 | 8-1 | X |
| 4 | 8-1 | X, P |
| 5 | 8-2 | X |
| 6 | 8-2 | X, P |
\X digest reactions
- 19.5 µL DNA
- 2.25 µL H2O
- 2.5 µL NEBuffer 2
- 0.5 µL XbaI
- 0.25 µL 100x BSA
\X digest master
- 9 µL H2O
- 10 µL NEBuffer 2
- 2 µL XbaI
- 1 µL 100x BSA
Pipette 5.5 µL into each reaction.
\X-P digest reactions
- 19.5 µL DNA
- 1.75 µL H2O
- 2.5 µL NEBuffer 3
- 0.5 µL XbaI
- 0.5 µL PstI
- 0.25 µL 100x BSA
\X-P digest master
- 7 µL H2O
- 10 µL NEBuffer 3
- 2 µL XbaI
- 2 µL PstI
- 1 µL 100x BSA
Pipette 5.5 µL into each reaction.
Digested for 6 hours at 45C, set to finish around 2300 tonight.
Restreak and colony PCR
I restreaked, colony PCR'd, and inoculated 10 colonies from Nick's ligation #5 that he plated on 2006-8-24 (bottom of link). They are restreaked on two plates.
Ligation conditions:
KaiC Gel + Gel Backbone #5 II at 17C
Colony PCR reaction
- 8 uL PCR supermix
- 1 uL VF2 primer
- 1 uL VR primer
Colony PCR master mix
- 88 uL PCR supermix
- 11 uL VF2 primer
- 11 uL VR primer
Pipette 10 uL into each reaction.
Colony PCR schedule
- 95C for 15'00
- Do 30 times:
- 95C for 0'30
- 55C for 0'30
- 72C for 2'00
- 72C for 10'00
- 4C forever
E-gel image of colony PCRs
It looks like the colonies don't contain our plasmid. (How can you tell? - See Below)
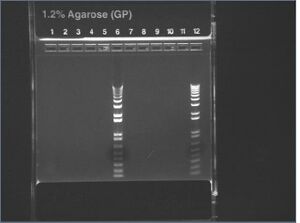
If there were no insert, we'd expect a 200bp band from a PCR. If there was no plasmid at all, our colonies would not have grown up under AMP selection. Ambiguously, no bands at all were seen.
E-gel image of \X and \X-P digests of Nick's KaiB + J04500 ligations
I forgot to run the undigested plasmid on this gel, but by comparing the \X and \X-P digests, it appears we still don't have insert. (How can you tell? - See Below) We expect ~530bp insert from this ligation.
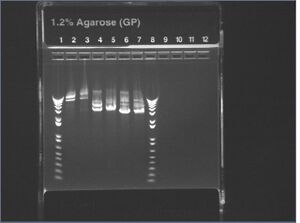
I'm having trouble interpreting this gel. The X-P cut should produce either a 530bp KaiB insert, a 200bp lac-rbs insert, or no insert at all. None of these options appear; we appear to get multiple 3+kb bands. Also, singly cut plasmid produces two bands, whereas doubly-cut plasmid produces only one band. It seems unclear how the presence or absence of any insert can be determined from this gel.