Haynes:cDNA Synthesis
Principle: In this procedure, complementary DNA (cDNA) is generated through a process called reverse transcription. It is similar to transcription except that the process uses mRNA as the template and DNA is the product. The process happens in vitro with components that are similar to a PCR reaction. Total mRNA (template) from cells is combined with dNTPs, MgCl2, primers called "oligo(dT)", a polymerase called Reverse Transcriptase, and buffer. Unlike mRNA, cDNA is very stable and can be quantified with PCR-based analyses. There are many kits on the market for cDNA synthesis. More kit-specific protocols will be added as needed.
SuperScript III
by Karmella Haynes, 2014
Materials
- Heat block set to 65°C
- Heat block set to 37°C
- Thermal Cycler (PCR machine)
- RNase-free 0.2 mL strip tubes, 0.5 mL tubes, 1.5 mL tubes
- Ice bucket (additional cold block is optional)
- SuperScript III First-Strand Synthesis kit (Life Technologies 18080-051)
- RNA samples - see the RNA extraction protocol
Procedure
cDNA SYNTHESIS
- If necessary, retrieve the RNA from the -80°C freezer and thaw on ice or a cold block. If freshly made, keep the samples on ice/ a cold block.
- NOTEBOOK ENTRY: Measure the concentration of all RNA samples (even if you have previously measured the RNA that was retrieved from the -80°C freezer. RNA degrades over time, even at -80°C).
- Retrieve the SuperScript III kit from the -20°C freezer.
- Thaw the following at room temperature on your bench: 50 μM oligo(dT) primer, 10 mM dNTP mix, Water, 10x RT buffer, 25 mM MgCl2, 0.1 M DTT
- Keep the following on ice or in a cold block: SuperScript III RT, RNaseOUT, RNase H.
- NOTEBOOK ENTRY: In labeled, clean RNase-free 0.5 mL tubes, set up oligo(dT) Primer-RNA annealing reactions.
- Incubate at 65°C/ 5 min. Immediately place on ice for 1 min.
- NOTEBOOK ENTRY: In a clean, RNase-free 1.5 mL tube, make enough cDNA synthesis mix for all desired reactions. Transfer 10 μL this mix into labeled, clean 0.2 mL, 8-tube PCR strip(s).
- Transfer each primer-RNA annealing reaction into a 10 μL aliquot of cDNA synthesis mix.
- Synthesize cDNA: Place the samples into the thermal cycler (PCR machine) and run the following program: 1x 50°C/ 50 min., 1x 80°C/ 5 min., 4°C/ ∞
- Degrade the RNA: Remove the samples from the thermal cycler. Add 1.0 μL RNase H to each sample. Mix by flicking the tubes and incubate at 37°C for 20 min.
- Proceed to stage B or store at -20°C. (Note: the cDNA is PCR-ready and does not need to be cleaned-up)
SAMPLE SET-UP FOR RT-PCR
HIGHLY recommended for organizing samples for downstream PCR analysis
- Get a fresh 8-tube PCR strip. These are MUCH easier to handle and store than individual 0.5 mL tubes.
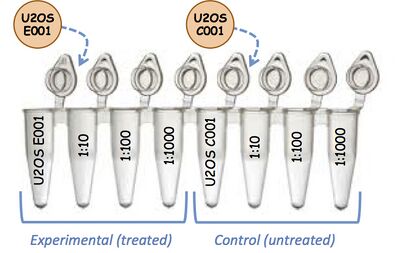
- Use four tubes for each unique cDNA sample. Label them in a fashion similar to the example illustration below.
- About this example: U2OS E001 is cDNA from a U2OS cell line that has been transfected with an experimental (E) transcription factor, while U2OS C001 represents the mock-transfected control (C) that was processed at the same time. Labels U2OS E002 and U2OS C002 will be used for the next experiment/ control set of cDNA. K562 E001/ C002 will be used when the cell type is changed to K562.
- General advice: You should use a labeling scheme that takes into account different cell types (if using different ones), and the number of cDNAs you will need to produce. For a time-course on a single cell type, a series of labels such as Ctrl 001, Dy2 001, Dy4 001, Dy6 001 could be used to label cDNAs from a 6-day-long, 2-day-interval experiment. The next time cDNA is made, the numbers could be switched to 002. The orange circles are stickers used to label the lid of the undiluted DNA. "1:10, 1:100, and 1:1000" designate the dilutions you will make.
- Transfer all 20 μL of cDNA sample 1 from the reaction tube into the first labeled tube (e.g., U2OS E001).
- Add the following volumes molecular biology-grade H2O to the next three tubes
- 1:10 = 90 μL
- 1:100 = 45 μL
- 1:1000 = 45 μL
- Transfer 10 μL cDNA into the 90 μL H2O in the "1:10" tube. Mix thoroughly by flicking the tube.
- Transfer 5 μL cDNA into the 45 μL H2O in the "1:100" tube. Mix thoroughly by flicking the tube.
- Transfer 5 μL cDNA into the 45 μL H2O in the "1:1000" tube. Mix thoroughly by flicking the tube.
- Repeat this process for all cDNA samples. There should be four tubes per cDNA sample: undiluted, 1:10 (for low-expressing genes), 1:100 (for intermediate-expressing genes), and 1:1000 (for highly-expressing genes like GAPDH, ACTB, and synthetic transgenes)
- Store all cDNA at -20°C.
Your NOTEBOOK ENTRIES should contain the following tables and information. Switch to edit mode on this page to copy-paste the code for the tables:
Measure RNA concentration
| Sample | OD 260 | 260/280 | ng/μL | Vol. to use for cDNA synth. |
| 1. ### | ### | ### | ### | ### μL (### μg) |
| 2. ### | ### | ### | ### | ### μL (### μg) |
oligo(dT) Primer-RNA annealing reactions
| Reagent | Vol |
| total RNA (up to 2μg) | up to 8 μL |
| 50 μM oligo(dT) primer | 1.0 |
| 10 mM dNTP mix | 1.0 |
| Water (SS III kit) | = 8.0 - vol. total RNA |
| 10.0 μL |
--> Incubate at 65°C/ 5 min. Immediately place on ice for 1 min.
cDNA synthesis mix
- Total reactions = N
- Samples:
- Names & description of cDNA sample 1
- Names & description of cDNA sample 2
- Names & description of cDNA sample 3
- etc.
| Reagent | Single rxn. | Mix (xN) |
| 10x RT buffer | 2.0 | ### |
| 25 mM MgCl2 | 4.0 | ### |
| 0.1 M DDT | 2.0 | ### |
| RNaseOUT | 1.0 | ### |
| SuperScript III RT | 1.0 | ### |
| 10.0 μL | ### μL |
--> Aliquot 10 μL of mix into 8-tube strip
--> Add annealing rxn. into each 10 μL aliquot
--> PCR machine: 50°C/ 50 min., 80°C/ 5 min., 4°C/ ∞
--> Add 1.0 μL RNase H, incubate at 37°C/ 20 min.
--> Store at -20°C