<- Back to Protocols
Type IIS Assembly
by Karmella Haynes, 2013
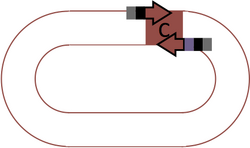
Principle: The familiar "BioBrick cloning" enzymes (i.e., EcoRI, NotI, XbaI, SpeI, PstI) are Type II restriction enzymes, which cut the sequences that they specifically bind to. The Type IIS Assembly method uses a Type IIS restriction enzyme, which binds at a specific sequence and cuts at a non-specific location exactly five base pairs away. As a result, the enzyme cleaves away its own binding site and leaves behind the most useful feature of assembly, sticky overhangs. When designed properly, Type IIS sites can be used to perform seamless assembly of parts. As an added convenience, this protocol allows cutting and ligation to occur in a single tube, as a single reaction. Thus, gel purification steps can be eliminated.
This protocol uses the Type IIS restriction enzyme BsmBI (CGTCTCn/nnnn).
| Use PCR to prepare the parts
|
- Multiple parts can be assembled in one step.
- Parts and the destination vector should be amplified by PCR.
- Make sure that none of the parts or the vector have any BsmBI sites!
|
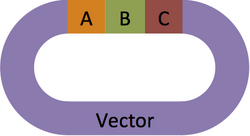 |
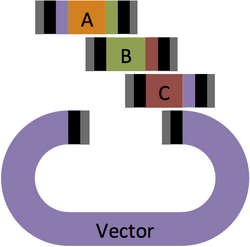
First, map out your assembly. In this example, three parts, A, B, and C will be assembled and inserted into a Vector.
|
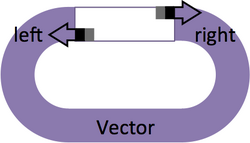 |
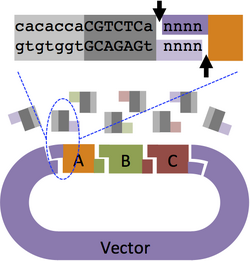
Design a pair of primers to add BsmBI sites to the ends of a vector backbone. The "cacacca" before BsmBI is used to help restriction enzyme positioning. The "a" after BsmBI is a spacer that is required to generate a correct 4-base sticky end.
Vector Primers
- Forward Primer: 5'-cacaccaCGTCTCa + 15 bp of "Vector right" top strand
- Reverse Primer: 5'-cacaccaCGTCTCa + 15 bp of "Vector left" bottom strand
pSB1A3 Vector Primers - already available in the Haynes lab freezer
- Forward Primer gg0001: 5'-cacaccaCGTCTCaactagtagcggccgctgcag
- Reverse Primer gg0002: 5'-cacaccaCGTCTCatctagaagcggccgcgaattcc
|
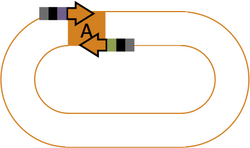 |
Part A Primers
- Forward Primer: 5'-cacaccaCGTCTCa + 4 bp of "Vector left" top strand + 15 bp of "Part A" top strand
Note: For insertion into pSB1A3, "4 bp of vector left" = TAGA
- Reverse Primer: 5'-cacaccaCGTCTCa + 4 bp of "Part B" bottom strand + 15 bp "Part A" bottom strand
|
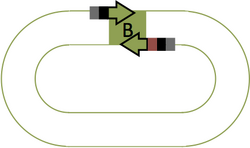 |
Part B Primers
- Forward Primer: 5'-cacaccaCGTCTCa + 15 bp of "Part B" top strand
- Reverse Primer: 5'-cacaccaCGTCTCa + 4 bp of "Part C" bottom strand + 15 bp "Part B" bottom strand
|
 |
Part C Primers
- Forward Primer: 5'-cacaccaCGTCTCa + 15 bp of "Part C" top strand
- Reverse Primer: 5'-cacaccaCGTCTCa + 4 bp of "Vector right" bottom strand + 15 bp "Part C" bottom strand
Note: For insertion into pSB1A3, "4 bp of Vector right bottom strand" = TAGT
|
 |
Run separate 50 μL PCR reactions for each part. If you are using plasmid DNA as a template, use no more than 10 ng in order to minimize carry-over into the final bacterial transformation step. Check 5 μL of the reaction on an agarose gel.
|
PCR Fragment prep
|
Digest the template DNA with DpnI
- Add 1 μL FastDigest DpnI and 5 μL of 10x FastDigest buffer to each PCR reaction. PCR product. Incubate at 37°C for 15 min. Note: DpnI cuts the methylated template DNA, not the unmethylated
| Reagent
|
Vol.
|
DpnI Digest
|
| PCR reaction |
45.0 μL
|
| 10x FD buffer (Fermentas) |
5.0
|
| DpnI (Fermentas) |
1.0
|
| dH2O |
---
|
| |
51.0 μL
|
- Purify the DNA from the 51 μL reaction using a Zymo clean and Concentrator kit (or similar PCR clean up kit).
Dilute the purified PCR product to 20 fmol/μL
- Measure ng/μL of the purified sample.
- The volume of purified DNA (x) you will need to dilute in a final volume of 20 μL = length in bp ÷ measured ng/μL * 20 fmols/μL * 650 fg/fmol ÷ 1,000,000 fg/ng * 20 μL final volume
- Formula: x = length in bp ÷ measured ng/μL * 0.013 * 20
- Note: final volume can be changed by changing the last number from 20 to something else.
|
| Digestion/ ligation reaction
|
Perform BsmBI/ T4 ligase mediated assembly
- BsmBI cuts the DNA fragments and creates complementary overhangs.
- Complementary sticky ends anneal via base pairing.
- T4 ligase seals gaps in the phosphodiester DNA backbone.
| Reagent
|
Vol.
|
Thermal cycling
- [45°C, 2 min.; 16°C 5 min.] x25
- 60°C, 10 min.
- 80°C, 20 min.
- 4°C, ∞
|
| 20 fmol of each DNA part |
up to 7.0 μL
|
| 10x T4 ligase buffer (Promega) |
1.0
|
| T4 ligase (NEB) |
1.0
|
| BsmBI |
0.5
|
| dH2O |
0.5
|
| |
10.0 μL
|
|
Bacterial transformation
- Add total volume (10.0 μL) to 50 μL chemically competent cells (e.g., BL21) in a 2.0 mL tube.
- Incubate on ice for 2 min., heat shock at 42°C for exactly 45 sec., immediately place on ice.
- Add 800 μL sterile SOC medium.
- Grow with shaking at 37°C for 30 min.
- Pellet the cells at top speed in a microcentrifuge for 3 min. at room temp.
- Discard the supernatant. Resuspend the cells in 100 μL LB + antibiotic.
- Plate cells on pre-warmed LB agar + antibiotic. Grow overnight at 37°C.
- Quick-transormation (e.g., DH5α-Turbo) is not recommended






