Haynes:SYBRGreen
From OpenWetWare
Jump to navigationJump to search
SYBER Green Assay: cDNA or genomic DNA
Workflow Overview
- Planning: Design your primers
- Planning: Design your reactions
- Wet bench: Make Gene Target (primer) master mixes
- Wet bench: Make Template (DNA) master mixes separately
- Wet bench: Add the appropriate combinations of of the master mixes to the 96-well plate
---> Run the reaction in the Light Cycler!
1. Design your primers
- Each gene/ locus you analyze requires a pair of oligos: forward primer and reverse primer
- The forward and reverse primers need to be ordered from a DNA synthesis company (e.g., IDT DNA, Promega, etc.).
- The primers should be between 100 - 150 bp apart to minimize differences in amplification efficiency.
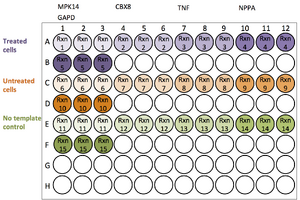
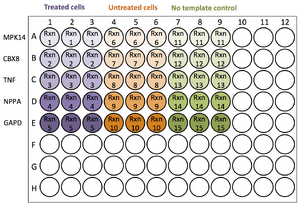
2. Set up a reaction list and plan the plate layout
- Determine how many wells and/or plates you will need. Each experimental cDNA or genomic DNA sample is a Template. The gene or locus being amplified is often referred to as a Gene Target. You should also include a reference Gene Target such as GAPD (a housekeeping gene that is always active, not expected to change). Each unique template and target combination requires its own reaction. You will also need to set up a no template control to observe the amount of background noise from the primers.
- Hypothetical example:
- Template DNA from treated cells, untreated cells, and a no template control = 3
- Four gene targets and a GAPD reference = 5
- Replicates per reaction = 3
- Wells needed = 3 templates * 5 targets * 3 replicates = 45 wells
- Conclusion: this experiment can be run in one 96-well plate
- A single plate contains 96 wells. To insure accuracy, three technical replicates per reaction (Rxn) are required
- If you need more than 96 wells, you must split the experiment over multiple plates.
- It is absolutely critical that you keep a reaction list and plate layout in your notes. Your plate set-up will probably vary for each run.
3. Reaction set-up: master mixes for each Gene Target
- Make ~750 nM F/R primer mixes: for each gene target, add 3.8 μL of 100 μM forward primer and 3.8 μL of 100 μM reverse primer to 492.4 μL H2O
- Label one 1.5 mL tube per gene target (final volume = 500 μL).
- Make enough PCR master mix for your plate...
- MPK14 is in 9 wells (Reactions 1, 6, and 11; 3 replicates each)
- Primer master mix amount = 9 + 0.5 to allow for pipetting error = x9.5
- Make primer master mixes for MPK14, CBX8, TNF, NPPA, and GAPD in separate tubes.
- Your notebook entry should include a list and table that are similar to the following:
Target Genes
- MPK14
- CBX8
- TNF
- NPPA
- GAPD (reference)
Primer master mixes (for this example, 5 tubes)
| Reagent | (Single well) | Gene Target (x9.5) |
| 2x SYBR Green Master | (7.5 μL) | 71.25 |
| 750 nM Forward/ Reverse primer mix | (3.0 μL) | 28.5 |
| Total vol. | (10.5 μL) | 99.75 |
4. Reaction set-up: master mixes for each Template
- For cDNA, make a 1:10 dilution of cDNA by adding 10 μL of the stock cDNA to 90 μL of PCR H2O.
- For cDNA in reference-gene reactions (e.g., GAPDH), make a 1:1000 dilution by adding 5 μL of 1:10 dilution to 45 μL of PCR H2O.
- For genomic DNA, the amount should be determined empirically. For your experiments, use the amount that produces a Ct or Cp value that is closest to 25 - 30 for the reference Target.
- Make enough Template master mix for your plate. For instance...
- Treated cells cDNA 1:10 is in 12 wells (Reactions 1, 2, 3, and 4; 3 replicates each)
- Template Master mix amount = 12 + 0.5 to allow for pipetting error = 12.5
- Treated cells cDNA 1:1000 is in 3 wells (Reaction 5; 3 replicates)
- Template Master mix amount = 3 + 0.5 to allow for pipetting error = 3.5
- Treated cells cDNA 1:10 is in 12 wells (Reactions 1, 2, 3, and 4; 3 replicates each)
- For cDNA, also make separate master mixes for "untreated cells 1:10," "untreated cells 1:1000," and "no template" in separate tubes.
- Your notebook entry should include a list and table that are similar to the following (this example is for cDNA):
Templates
- Treated cells 1:10
- Treated cells 1:1000
- Untreated cells 1:10
- Untreated cells 1:1000
- No-template control (PCR H2O)
Template master mixes (for this example, 5 tubes)
| Reagent | (Single well) | cDNA 1:10 (x12.5) | cDNA 1:1000 (x3.5) | no template (x15.5)* |
| cDNA or PCR H2O | (2.0 μL) | 25.0 | 7.0 | 31.0 |
| PCR H2O | (2.5 μL) | 31.25 | 8.75 | 38.75 |
| Total vol. | (4.5 μL) | 56.25 | 15.75 | 69.75 |
*For the no template control, use PCR H2O instead of cDNA.
Important notes about Genomic DNA templates
- Genomic DNA templates tend to be more diluted (fewer per cell) compared to cDNA because genomic loci exist as one copy per haploid genome, whereas mRNAs (and the cDNA produced from them) exist as multiple copies per gene (depending upon expression level).
- Genomic DNA - We recommend 10 ng DNA/ 15 μL rxn. One human cell has ~7 pg DNA, so 10 ng (10,000 pg) is about 1,429 cell genomes, a good sample size for polymorphism ratio studies. Theoretically, a genomic DNA prep from 1 million cells (1 well from a 6-well dish), should yield 7 μg DNA, which would be 70 ng/μL if suspended in a 100 μL volume.
- Sheared ChIP DNA - The recommended volume for ChIP DNA is 2.0 μL. If you observe very weak signal (high Ct or Cp) from 2.0 μL of undiluted genomic DNA template, increase the volume of template and decrease the volume of PCR H2O. You can use of to 6.5 μL of template, and as low as zero PCR H2O.
5. Reaction set-up: loading the 96-well plate
The following illustrations use Plate Layout Variation 2...
| Pipette 19.5 μL of Template master mixes into the first well of each 3-well group |
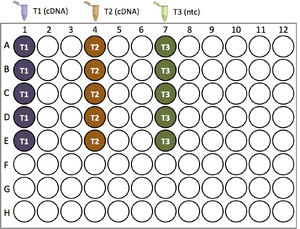
|
| Add 25.5 μL of Gene Target master mix to the cDNA. After each addition, mix by gently pipetting up and down 3 - 5 times (without making bubbles). | 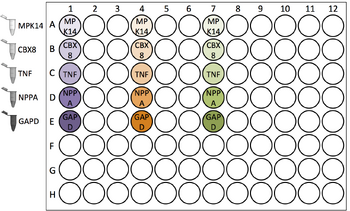
|
| Transfer 15 μL of solution from A1 into A2, and A3. Use a fresh pipette tip to do the same for A4-6, and A7-8. | 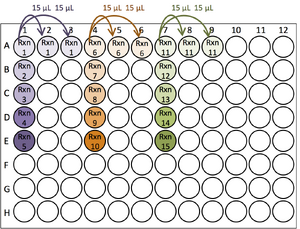
|
| Repeat this procedure for the rest of the plate | 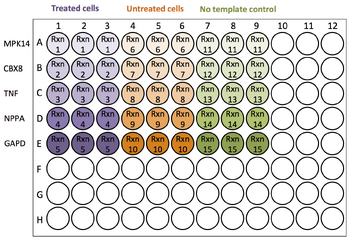
|
- Seal the plate with clear film.
- The plate is now ready to load into the Light Cycler 480!