Gill:Spot complementation test
Rev. 2015-03-05
Adapted from: Hendrickson & McCorquodale 1971, Genetic and Physiological Studies of Bacteriophage T5: I. An Expanded Genetic Map of T5. J Virol 7: 612-618, PMID 16789131.
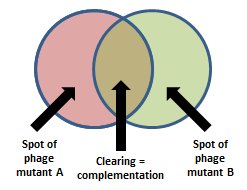
The purpose of this assay is to determine if mutations in a set of phages are present in the same gene or in different genes; the phages are tested pairwise. Host cells that are not permissive for phage growth are co-infected with two different phage mutants. If the two phages have defects in different essential genes, then the co-infected cell will contain one functional copy of each gene, and this in trans complementation will allow the phages to lyse the cell and form a plaque. If the two phages have defects in the same gene, they will not be able to complement and no plaques will form.
Materials
- Agar plates
- T-top agar (in 4 ml aliquots at 48-50 ºC)
- Fresh overnight or over-day culture of non-permissive host cells
- Stocks of phage mutants, adjusted to ~107 PFU/ml in lambda diluent or LB
Procedure
- Label the bottom(s) of the plates with each pairwise combination of phage you will test. You should be able to fit 6 phage pairs on each plate.
- Add 100 µl of the bacterial culture to a 4 ml T-top aliquot, mix and pour over the surface of a plate. Allow the lawn to set for a few minutes.
- Apply a 10 µl drop of one phage from each test pair to the plate, allow the plate to air-dry for ~10 min or until the drop is adsorbed into the lawn.
- Apply a 10 µl drop of the second phage from each test pair to the plate to form two partially overlapping spots for each pair. Allow the plate to air-dry for ~10 min or until the second drop is adsorbed into the lawn.
- Incubate the plates overnight at 37 ºC and observe the plates for clearing. Clearing in the overlap region where cells were mixedly infected indicates complementation.
Controls: spot 10 µl of the wt phage to a lawn as a positive control, and spot each phage against itself as a negative control.
Notes
Please feel free to post comments, questions, or improvements to this protocol. Happy to have your input!