GFP
From OpenWetWare
Jump to navigationJump to search
General Info
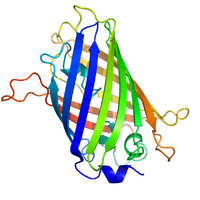
- Originally discovered and characterized in the jellyfish Aeqorea victoria
- wtGFP sequence on NCBI
- 238 amino acids
- The tertiary structure is a fluorophore [Ser(65)-Tyr(66)-Gly(67)] nestled inside a protective beta barrel.
- Oxygen is required for maturation of the fluorophore
- Natural state is a dimer with a Kd of ~ 100 µM.
GFP Variants
- GFPmut2 - S65A, V68L, S72A [1]
- GFPmut3b - S65G, S72A [1]
- GFPmut3* - mutations from wt: S2R,S65G,S72A
- This is GFPmut3b with an accidental mutation at position 2 that doesn't affect function according to the authors.[2]
- Emerald - F64L, S65T, S72A, N149K, M153T, I167T
- EGFP - inserted GTG as second codon, F64L, S65T + optimized for human codons with 35-fold increase in fluorescence over GFP [3]
- Additionally, "mutation of His231 to Leu, which was probably inadvertent and neutral". [3]
- yemGFP - F64L, S65T, A206K [4]
- superfolder GFP - S30R, Y39N, F64L, (S65T/G65T), F99S, N105T, Y145F, M153T, V163A, I171V, A206V [5]
- Inherited parent variant mutations (GFPmut3*): S2R,
S65G, S72A (S65G mutation is superseded by S65T mutation, hence the alternative mutation designation of G65T)
- Inherited parent variant mutations (GFPmut3*): S2R,
- Other variants and their mutations can be found in Shaner supplementary table 2 [6]
Mutations
- F64L: improve folding @37C
- S65T: 5-6x increase in amplitude and red shift
- S65(G|T) and T203(Y|F|W|H): GFP -> YFP
- Y66W: GFP -> CFP
- Y66H: GFP -> BFP (more blue than CFP), dim, easily photobleached
- Y66F: excitation 360nm, emission 442nm
- R96A: slows cyclization reaction from minutes to months
- F99S, M153T, V163A: "cycle 3" mutations (originally reported as F100S, M154T, V164A) [7]
- Y203I: eliminate excitation peak at 475nm, leaving lower peak of 399nm. emission remains at 511nm, producing large Stokes shift
- A206K: make monomeric
- S30R, Y39N, N105T, Y145F, I171V, A206V: additional superfolder mutations [5]
References
General
- Cormack BP, Valdivia RH, and Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP). Gene. 1996;173(1 Spec No):33-8. DOI:10.1016/0378-1119(95)00685-0 |
- Andersen JB, Sternberg C, Poulsen LK, Bjorn SP, Givskov M, and Molin S. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl Environ Microbiol. 1998 Jun;64(6):2240-6. DOI:10.1128/AEM.64.6.2240-2246.1998 |
- Li X, Zhang G, Ngo N, Zhao X, Kain SR, and Huang CC. Deletions of the Aequorea victoria green fluorescent protein define the minimal domain required for fluorescence. J Biol Chem. 1997 Nov 7;272(45):28545-9. DOI:10.1074/jbc.272.45.28545 |
- Stricker J, Cookson S, Bennett MR, Mather WH, Tsimring LS, and Hasty J. A fast, robust and tunable synthetic gene oscillator. Nature. 2008 Nov 27;456(7221):516-9. DOI:10.1038/nature07389 |
- Pédelacq JD, Cabantous S, Tran T, Terwilliger TC, and Waldo GS. Engineering and characterization of a superfolder green fluorescent protein. Nat Biotechnol. 2006 Jan;24(1):79-88. DOI:10.1038/nbt1172 |
- Shaner NC, Steinbach PA, and Tsien RY. A guide to choosing fluorescent proteins. Nat Methods. 2005 Dec;2(12):905-9. DOI:10.1038/nmeth819 |
- Crameri A, Whitehorn EA, Tate E, and Stemmer WP. Improved green fluorescent protein by molecular evolution using DNA shuffling. Nat Biotechnol. 1996 Mar;14(3):315-9. DOI:10.1038/nbt0396-315 |
- Prasher DC, Eckenrode VK, Ward WW, Prendergast FG, and Cormier MJ. Primary structure of the Aequorea victoria green-fluorescent protein. Gene. 1992 Feb 15;111(2):229-33. DOI:10.1016/0378-1119(92)90691-h |
original cloning of GFP
- Chalfie M, Tu Y, Euskirchen G, Ward WW, and Prasher DC. Green fluorescent protein as a marker for gene expression. Science. 1994 Feb 11;263(5148):802-5. DOI:10.1126/science.8303295 |
original use of GFP as a reporter
- Tsien RY. The green fluorescent protein. Annu Rev Biochem. 1998;67:509-44. DOI:10.1146/annurev.biochem.67.1.509 |
good review of GFP
-
Molecular Biology and Muation of Green Fluorescent Protein (Book Chapter, Zacharias & Tsien)
GFP as a measure of gene expression
- Leveau JH and Lindow SE. Predictive and interpretive simulation of green fluorescent protein expression in reporter bacteria. J Bacteriol. 2001 Dec;183(23):6752-62. DOI:10.1128/JB.183.23.6752-6762.2001 |
- Iafolla MA, Mazumder M, Sardana V, Velauthapillai T, Pannu K, and McMillen DR. Dark proteins: effect of inclusion body formation on quantification of protein expression. Proteins. 2008 Sep;72(4):1233-42. DOI:10.1002/prot.22024 |
- Bagh S, Mazumder M, Velauthapillai T, Sardana V, Dong GQ, Movva AB, Lim LH, and McMillen DR. Plasmid-borne prokaryotic gene expression: sources of variability and quantitative system characterization. Phys Rev E Stat Nonlin Soft Matter Phys. 2008 Feb;77(2 Pt 1):021919. DOI:10.1103/PhysRevE.77.021919 |
- Dong GQ and McMillen DR. Effects of protein maturation on the noise in gene expression. Phys Rev E Stat Nonlin Soft Matter Phys. 2008 Feb;77(2 Pt 1):021908. DOI:10.1103/PhysRevE.77.021908 |