Free Genes/FreeGenes MoClo
In order to realize a future where hundreds or thousands of people can share DNA easily and quickly, we believe that the way in which DNA is assembled should be standardized and modular. For this reason, we have developed a backward-compatible MoClo standard to use with all Free Gene Project genes.
MoClo Introduction
MoClo cloning is a cloning method developed by Ernst Weber and Carola Engler in 2011 to faciliate scalable modular assembly of DNA [1]. Several groups [2][3][4][5] have made toolkits in order to work with different organisms, while still retaining the same assembly method. t is even accepted at iGem as an assembly standard that can be deposited [6].
The MoClo assembly method can assembly over 10 parts at once in a one-pot reaction[7], making it well suited for assembling entire genetic circuits in one or two reactions.
Basics of MoClo
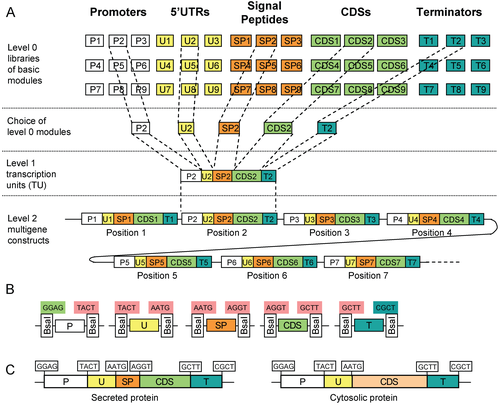
MoClo assembly is a subset of GoldenGate assembly, which uses Type IIS restriction enzymes. Type IIS restriction enzymes cut outside of their recognition site, and this special property allows for many different parts to be assembled at once with fewer restriction enzymes[8].
GoldenGate assembly typically uses a restriction enzyme, such as BsaI in combination with a ligase, such as T4 ligase, in a single one-pot reaction. All DNA parts are added to the mixture, and using predefined overlaps, are stitched together. This reaction is extremely efficient, and results in DNA that is ready to transform without any downstream preparation.
MoClo expands upon GoldenGate assembly by simply defining enzymes and overlaps. In its most basic definition, MoClo parts are flanked by BsaI with unique cut sites. For example, CDSs are flanked with AATG (with the ATG being part of the start codon) and GCTT. This allows for anyone with a CDS to combine that CDS with any combination of promoter or terminator, allowing for scalable redistribution of DNA.
Problems with MoClo
MoClo has several problems which have manifested themselves in toolkits. In particular, there was no standardization of tagging for proteins, on either the 3' or 5' ends. This has caused for there to be many alternative definitions[1][7][9] of the relationship of tags to the CDS parts.
Another major problem is the divide between communities. Since generally communities that work with certain organisms tend to not have to collaborate with outside communities, this can create an individual environment which breaks the definition[7] [10].
A better method would retain backwards compatibility to all the other MoClo methods, have better ways to tag proteins, and be maintained by an organization who is agnostic about organism choice. For these reasons, we began the development of the FreeGenes MoClo method.
FreeGenes MoClo
The FreeGenes MoClo method was developed for the FreeGenes project [11]. This cloning method uses a couple of concepts derived from the Yeast Toolkit[7], while expanding upon a couple of other areas.
- FreeGenes definitions
- Part
- A piece of DNA that is flanked by MoClo overhangs.
- Overhang
- A 4 base pair sequence that is cut by an enzyme. Flanks a part. These are defined by short 1-2 letter names, which correspond to a 4 base pair sequence.
- Primitive
- A part that is not derived from other parts. These can be created by synthesis, PCR, or possibly even restriction enzyme cloning.
- Composite
- A part that is derived from other parts.
- Linker
- The flanking sequence between the vector and parts. Defines the overhangs for a new composite.
- Vector
- The backbone plasmid DNA that carries parts. Originally carries negative selection marker, which is replaced by a part or a collection of parts and 2 linkers. Afterwards, it can be cloned out of, using overhangs defined by the linkers.
- Object
- The entire plasmid that carries a part, including the vector, linkers, and part(s). Usually this describes a physical molecule.
FreeGenes Primitives
A FreeGene primitive is a part that is compatible with MoClo assembly. It connects with other primitives into a composite part. Below is a few simple examples of primitive part types that can be synthesized through the FreeGenes Project.
| Part Type | overhangs | prefix | suffix |
| prokaryotic promoter | X-i0 | GGAG | TACT |
| eukaryotic promoter | X-i1 | GGAG | AATG |
| RBS | i0-i1 | TACT | AATG |
| CDS | i1-i2 | AATG | GCTT |
| terminator | i2-Y | GCTT | CTAG |
CDS primitive
The CDS primitive is unique in a few optimizations that are on the 3' end in order to allow for adding tags in a seamless way. The last coding codon is standardized to 1 of 20 codons. The stop codon is standardized to TGA. This results in the following configuration:
NNN TGA
A small insert sequence (AGA) and the MoClo overhang (GCTT) is then added to the end.
NNN TGA AGA GCTT
This sequence is equivalent to
NNNT GAAGAGC TT
Which includes the reverse complement to the SapI recognition sequence (GCTCTTCN). That means SapI cuts the last codon (NNN) of any protein.
NNN TGAAGAGCTT
This allows for any protein sequence to be cut with SapI. The last codon is standardized according to the following table.
| Amino acid | Codon |
| M | ATG |
| W | TGG |
| F | TTT |
| L | CTG |
| I | ATT |
| V | GTG |
| S | TCC |
| P | CCA |
| T | ACC |
| A | GCC |
| Y | TAC |
| H | CAT |
| Q | CAG |
| N | AAC |
| K | AAG |
| D | GAT |
| E | GAG |
| C | TGC |
| R | CGC |
| G | GGC |
FreeGenes Overhangs
FreeGenes defines a few overhangs, listed below. X - Y flank parts, A - B flank vector, and the region between A - X and Y - B are defined to be linker sequences. Unless otherwise noted, all overhangs are cut with BsaI.
| X | GGAG |
| i0 | TACT |
| i1 | AATG |
| i2 | GCTT |
| Y | CGCT |
| A | ATCC |
| v0 | GTGA |
| v1 | AGGA |
| v2 | CTTC |
| v3 | CCAG |
| B | TCGG |
| v4 | TAGC |
| S | ACTC |
Part Types
References
- ↑ 1.0 1.1 1.2 Ernst Weber , Carola Engler , Ramona Gruetzner, Stefan Werner, Sylvestre Marillonnet (2011-02-18). "A Modular Cloning System for Standardized Assembly of Multigene Constructs". PLOS ONE. [1]
- ↑ Engler, Youles M, Gruetzner R, Ehnert TM, Werner S, Jones JD, Patron NJ, Marillonnet S. (2014-11-21). "A golden gate modular cloning toolbox for plants." ACS Synthetic Biology. [2]
- ↑ Sonya V. Iverson, Traci L. Haddock, Jacob Beal, and Douglas M. Densmore (2015-10-19). "CIDAR MoClo: Improved MoClo Assembly Standard and New E. coli Part Library Enable Rapid Combinatorial Design for Synthetic and Traditional Biology". ACS Synthetic Biology. [3]
- ↑ Simon J. Moore†, Hung-En Lai†, Richard J. R. Kelwick†, Soo Mei Chee, David J. Bell, Karen Marie Polizzi, Paul S. Freemont (2016-04-20). "EcoFlex: A Multifunctional MoClo Kit for E. coli Synthetic Biology". ACS Synthetic Biology. [4]
- ↑ Niels Wicke, David Radford, Valeria Verrone, Anil Wipat, Christopher E. French (2017-09-06) "BacilloFlex: A modular DNA assembly toolkit for Bacillus subtilis synthetic biology". bioRxiv. [5]
- ↑ iGem (2016). "What are PhytoBricks?". iGem. [6]
- ↑ 7.0 7.1 7.2 7.3 Michael E. Lee†, William C. DeLoache, Bernardo Cervantes, John E. Dueber (2015-04-15). "A Highly Characterized Yeast Toolkit for Modular, Multipart Assembly". ACS Synthetic Biology. [7]
- ↑ NEB. "Golden Gate Assembly" NEB. [8]
- ↑ Nicola J. Patron et al (2015-07-14). "Standards for plant synthetic biology: a common syntax for exchange of DNA parts". New Phytologist. [9]
- ↑ Ulrike Obst†, Timothy K. Lu, Volker Sieber (2017-03-02). "A Modular Toolkit for Generating Pichia pastoris Secretion Libraries." ACS Synthetic Biology. [10]
- ↑ BioBricks Foundation. "The Free Genes Project". biobricks. [11]