Forster Lab
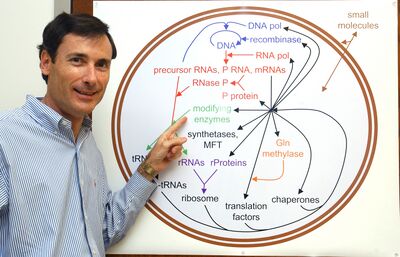
The Boss!?
Professor Anthony C. Forster, M.D., Ph.D.
Head, Program in Molecular Biology
Department of Cell and Molecular Biology
Uppsala University
office and mailing address:
ICM Dept., Room D9:216b
Husargatan 3, Box 596
75124 Uppsala, Sweden
office and cell phone: +46-18-471 4618
e-mail: a.forster(at)icm.uu.se
lab web: http://openwetware.org/wiki/Forster_Lab
Department web: http://www.icm.uu.se/
Uppsala University web: http://www.uu.se/en/
Biography:
Anthony C. Forster (Ph.D. Biochem., U. Adelaide; M.D., Harvard U.) researches RNA, protein synthesis and applications thereof (synthetic biology). He discovered the hammerhead catalytic RNA structure, invented external guide sequences for ribonuclease P, and created unnatural genetic codes de novo, all of which founded biotech companies. He has published in journals including Cell, Nature and Science, edited volumes of Methods and Biotechnology J., and coauthored "Synthetic Biology: A Lab Manual."
Research Description:
Synthetic biology and protein synthesis
SynBio is a creative new field defined as the complex engineering of replicating systems. It encompasses next-generation technology for bioengineering and fresh approaches to global challenges such as drug discovery and biofuels.
Our current projects include:
1. Improving ribosomal incorporation of unnatural amino acids for investigating translation mechanism and for applications such as directed evolution of peptidomimetic drugs.
2. Determining functions of ribosomal RNA modifications and ribosomal proteins towards synthesis of the ribosome and self-replication.
3. Understanding and improving transcription termination.
4. Improving vaccines via synthetic biology.
Research Keywords:
Synthetic biology, protein synthesis, drug discovery, transcription termination, chromoprotein, fluorescent protein, unnatural amino acid, directed evolution, translation, ribosome, RNA, modification enzyme, E. coli, bacteria, microbiology, biochemistry, vaccine, Tony Forster
Meet our lab group:

phones in lab:
+46-18-471 4387, 471 4651 and 471 4204
Teaching:
1MB433: Synthetic Biology (10 hp, weeks 4-9)
http://www.uu.se/en/education/master/selma/kursplan/?kpid=24757&type=1
1BG044: Frontiers in Bioscience (10 hp, weeks 37-51)
https://www.uu.se/en/admissions/master/selma/kursplan/?kpid=39610&type=1
1MB205 and 1MB405 Project in laboratory synthetic biology I and II (Uppsala iGEM) courses, co-director
iGEM 2020 Uppsala University Team Members:
https://2020.igem.org/Team:UofUppsala/Team
Congratulations on winning "Best new application" prize!
iGEM 2014 Ravenwood High School Team Members:
http://2014hs.igem.org/Team:Ravenwood_Raptors
Congratulations on being the first iGEM team from Tennessee!

http://www.worldscientific.com/worldscibooks/10.1142/9061#t=aboutBook
Postdoctoral Applicants:
Minimal qualifications include expertise in molecular biology and 2 first-authored research papers in international peer-reviewed journals. Experience with bacterial translation or RNA is a plus. Please mail a letter of interest and C.V.
Master and PhD Applicants:
Students enrolled in a Master program (including EU students seeking an Erasmus exchange) are welcome to apply for a few months' work experience in the lab. Most students achieve authorship on a publication, and extension into a PhD position is possible. Research experience is a plus. Please mail a letter of interest and C.V.
PATENTS:
Forster, A C, Blacklow, S C. Process and compositions for peptide, protein and peptidomimetic synthesis. US6977150
(founding I.P. for Ra Pharmaceuticals, Inc., Boston, now merged with UCB).
Altman, S, Forster, A C, Guerrier-Takada, C L. Cleavage of targeted RNA by RNAase P. US5168053
(founding I.P. for Innovir Laboratories, Inc., NY).
PUBLICATIONS:
Almost all pubs indexed by and available from PubMed:
http://www.ncbi.nlm.nih.gov/pubmed/ (type "Forster AC")
Citations per paper are available from Google Scholar:
http://scholar.google.com/ (type "Forster AC")
Forster, A C, Bao, L, Liljeruhm, J. Synthetic biology: A lab manual. Second edition. World Scientific Press, in press
Bao, L, Liljeruhm, J, Crespo Blanco, R, Brandis, G, Remme, J, Forster, A C. Translational impacts of enzymes that modify
ribosomal RNA around the peptidyl transferase centre. RNA Biol., 21, 31-41, 2024
Open access: https://doi.org/10.1080/15476286.2024.2368305
Bao, L, Karpenko, V V, Forster, A C. Rate-limiting hydrolysis in ribosomal release reactions revealed by ester activation. J. Biol. Chem., 298, 102509, 1-9, 2022
Open access: https://doi.org/10.1016/j.jbc.2022.102509
Forster, A C. Tales of the unexpected in Sidney Altman's laboratory. RNA, 28, 1406-1408, 2022
Open access: doi: 10.1261/rna.079397.122
https://rnajournal.cshlp.org/content/early/2022/09/16/rna.079397.122.short
Liljeruhm, J, Leppik, M, Bao, L, Truu, T, Calvo-Noriega, M, Freyer, N S, Liiv, A, Wang, J, Crespo Blanco, R, Ero,
R, Remme, J, Forster, A C. Plasticity and conditional essentiality of modification enzymes for domain V of Escherichia coli 23S ribosomal RNA. RNA, 28, 796-807, 2022
doi: 10.1261/rna.079096.121
Forster, A C. Revisiting the extinction of the RNA world. Biochemistry, 61, 749-751, 2022
Open access https://doi.org/10.1021/acs.biochem.2c00121
Doerr, A, Foschepoth, D, Forster, A C, Danelon, C. In vitro synthesis of 32 translation-factor proteins from a single template reveals impaired ribosomal processivity. Sci. Rep. 11:1898, 1-12, 2021
Open access https://doi.org/10.1038/s41598-020-80827-8
Bao, L, Menon, P N K, Liljeruhm, J, Forster, A C. Overcoming chromoprotein limitations by engineering a red fluorescent protein. Anal. Biochem. 611:113936, 1-8, 2020
Open access https://doi.org/10.1016/j.ab.2020.113936
Vogel, C, Gynnå, A, Yuan, J, Bao, L, Liljeruhm, J, Forster, A C. Rationally-designed Spot 42 RNAs with an inhibition/toxicity profile advantageous for engineering E. coli. Engineering Rep., 2:3, e12126, 1-10, 2020
Our front cover and open access: https://doi.org/10.1002/eng2.12126
Liljeruhm, J, Wang, J, Kwiatkowski, M, Sabari, S, Forster, A C. Kinetics of D-amino acid incorporation in translation. ACS Chem. Biol. 14:204-213, 2019
Open access: https://doi.org/10.1021/acschembio.8b00952
Wang, J, Forster, A C. Ribosomal incorporation of unnatural amino acids: lessons and improvements from fast kinetics studies. Curr. Opin. Chem. Biol. 46:180-187, 2018
Open access: https://doi.org/10.1016/j.cbpa.2018.07.009
Liljeruhm, J, Funk, S K, Tietscher, S, Edlund, A D, Jamal, S, Wistrand-Yuen, P, Dyrhage, K, Gynnå, A, Ivermark, K, Lövgren, J, Törnblom, V, Virtanen, A, Lundin, E R, Wistrand-Yuen, E, Forster, A C. Engineering a palette of eukaryotic chromoproteins for bacterial synthetic biology. J. Biol. Eng. 12:8, 1-10, 2018
https://doi.org/10.1186/s13036-018-0100-0
Shepherd, T R, Du, L, Liljeruhm, J, Samudyata, Wang, J, Sjödin, M O D, Wetterhall, M, Yomo, T, Forster, A C. De novo design and synthesis of a 30-cistron translation-factor module. Nucleic Acids Res. 45, 10895-10905, 2017
Wang, J, Forster, A C. Translational roles of the C75 2'OH in an in vitro tRNA transcript at the ribosomal A, P and E sites. Sci. Rep. 7, 6709, 1-8, 2017
Wang, J, Kwiatkowski, M, Forster, A C. Ribosomal peptide syntheses from activated substrates reveal rate limitation by an unexpected step at the peptidyl site. J. Am. Chem. Soc. 138, 15587-15595, 2016
Wang, J, Kwiatkowski, M, Forster, A C. Kinetics of tRNAPyl-mediated amber suppression in E. coli translation reveals unexpected limiting steps and competing reactions. Biotechnol. Bioeng. 113, 1552-1559, 2016
Wang, J, Kwiatkowski, M, Forster, A C. Kinetics of ribosome-catalyzed polymerization using artificial aminoacyl-tRNA substrates clarifies inefficiencies and improvements. ACS Chem. Biol. 10, 2187-2192, 2015
Kwiatkowski, M, Wang, J, Forster, A C. Facile synthesis of N-acyl-aminoacyl-pCpA for preparation of mischarged fully ribo tRNA. Bioconjugate Chem. 25, 2086-2091, 2014
Liljeruhm, J, Gullberg, E, Forster A C. Synthetic biology: A lab manual. World Scientific Press, 204 pp, 2014
http://www.worldscientific.com/worldscibooks/10.1142/9061#t=aboutBook
Wang, J, Kwiatkowski, M, Pavlov, M Y, Ehrenberg, M, Forster, A C. Peptide formation by N-methyl amino acids in translation is hastened by higher pH and tRNAPro. ACS Chem. Biol. 9, 1303-1311 and front cover, 2014
Ieong, K-W, Pavlov, M Y, Kwiatkowski, M, Ehrenberg, M, Forster, A C. A tRNA body with high affinity for EF-Tu hastens ribosomal incorporation of unnatural amino acids. RNA 20, 632-643, 2014
Punekar, A, Liljeruhm, J, Shepherd, T R, Forster, A C, Selmer, M. Structural and functional insights into the molecular mechanism of rRNA m6A methyltransferase RlmJ. Nucleic Acids Res. 41, 9537-9548, 2013
Quax, T E F, Wolf, Y I, Koehorst, J J, Wurtzel, O, van der Oost, R, Ran, W, Blombach, F, Makarova, K S, Brouns, S J J, Forster, A C, Wagner, E G H, Sorek, R, Koonin, E V, van der Oost, J. Differential translation tunes uneven production of operon-encoded proteins. Cell Rep. 4, 938-944, 2013
Ieong, K-W, Pavlov, M Y, Kwiatkowski, M, Forster, A C, Ehrenberg, M. Inefficient delivery but fast peptide bond formation of unnatural L-aminoacyl-tRNAs in translation, J. Am. Chem. Soc. 134, 17955-17962, 2012
Punekar, A, Shepherd, T R, Liljeruhm, J, Forster, A C, Selmer, M. Crystal structure of RlmM, the 2'O-ribose methyltransferase for C2498 of E. coli 23S rRNA. Nucleic Acids Res., 40, 10507-10520, 2012
Forster, A C. Synthetic biology challenges long-held hypotheses in translation, codon bias and transcription. Biotech. J., 7, 835-845, 2012
Forster, A C, Lee, S Y. Editorial: NextGen SynBio has arrived... Biotech. J., 7, 827, 2012
Du, L, Villarreal, S, Forster, A C. Multigene expression in vivo: supremacy of large versus small terminators for T7 RNA polymerase. Biotechnol. Bioeng., 109, 1043-1050, 2012
Wang, H H, Huang, P-Y, Xu, G, Haas, W, Marblestone, A, Li, J, Gygi, S P, Forster, A C, Jewett, M C, Church, G M. Multiplexed in vivo His-tagging of enzyme pathways for in vitro single-pot multienzyme catalysis. ACS Synth. Biol., 1, 43-52, 2012
Watts, R E, Forster, A C. Update on pure translation display with unnatural amino acid incorporation. Meth. Mol. Biol., 805, 349-365, 2012
Gao, R, Forster, A C. Changeability of individual domains of an aminoacyl-tRNA in polymerization by the ribosome. FEBS Lett., 584(1), 99-105, 2010
Jewett, M C, Forster, A C. Update on designing and building minimal cells. Curr. Opin. Biotech., 21, 697-703, 2010
Watts, R E, Forster, A C. Chemical models of peptide formation in translation. Biochem., 49, 2177-2185, 2010
Du, L, Gao, R, Forster, A C. Engineering multigene expression in vitro and in vivo with small terminators for T7 RNA polymerase. Biotechnol. Bioeng., 104, 1189-1196, 2009
Forster, A C. Low modularity of aminoacyl-tRNA substrates in polymerization by the ribosome. Nucleic Acids Res., 37, 3747-3755, 2009
Pavlov, M Y, Watts, R E, Tan, Z, Cornish, V W, Ehrenberg, M, Forster, A C. Slow peptide bond formation by proline and other N-alkylamino acids in translation. Proc. Natl. Acad. Sci. USA, 106(1), 50-4, 2009
Forster, A C, Church, G M. Synthetic biology projects in vitro. Genome Res., 17(1), 1-6 and front cover, 2007
Zhang, B, Tan, Z, Gartenmann Dickson, L, Nalam, M N L, Cornish, V W, Forster, A C. Specificity of Translation for N-Alkyl Amino Acids. J. Am. Chem. Soc., 129(37), 11316-11317, 2007
Forster, A C, Church, G M. Towards synthesis of a minimal cell. Mol. Syst. Biol., 2(45), 1-10, 2006
Forster, A C. Engineering translation: A nano-review. Methods, 36(3), 225-6, 2005
Tan, Z, Blacklow, S C, Cornish, V W, Forster, A C. De novo genetic codes and pure translation display. Methods, 36(3), 279-90, 2005
Forster, A C, Cornish, V W, Blacklow, S C. Pure translation display. Anal. Biochem., 333(2), 358-64, 2004
Tan, Z, Forster, A C, Blacklow, S C, Cornish, V W. Amino acid backbone specificity of the Escherichia coli translation machinery. J. Am. Chem. Soc., 126(40), 12752-3, 2004
Forster, A C, Tan, Z, Nalam, M N L, Lin, H, Qu, H, Cornish, V W, Blacklow, S C. Programming peptidomimetic syntheses by translating genetic codes designed de novo. Proc. Natl. Acad. Sci. USA, 100(11), 6353-7, 2003
Forster, A C, Weissbach, H, Blacklow, S C. A simplified reconstitution of mRNA-directed peptide synthesis: activity of the epsilon enhancer and an unnatural amino acid. Anal. Biochem., 297(1), 60-70, 2001
Li, E, Beard, C, Forster, A C, Bestor, T H, Jaenisch, R. DNA methylation, genomic imprinting, and mammalian development. Cold Spring Harb. Symp. Quant. Biol., 58, 297-305, 1993
Forster, A C, Altman, S. External guide sequences for an RNA enzyme. Science, 249(4970), 783-6, 1990
Forster, A C, Altman, S. Similar cage-shaped structures for the RNA components of all ribonuclease P and ribonuclease MRP enzymes. Cell, 62(3), 407-9, 1990
Forster, A C, Davies, C, Hutchins, C J, Symons, R H. Characterization of self-cleavage of viroid and virusoid RNAs. Meth. Enzymol., 181, 583-607, 1990
McInnes, J L, Forster, A C, Skingle, D C, Symons, R H. Preparation and uses of photobiotin. Meth. Enzymol., 184, 588-600, 1990
Forster, A C, Davies, C, Sheldon, C C, Jeffries, A C, Symons, R H. Self-cleaving viroid and newt RNAs may only be active as dimers. Nature, 334(6179), 265-7, 1988
McInnes, J L, Forster, A C, Symons, R H. Photobiotin-labelled DNA and RNA hybridization probes. Meth. Mol. Biol., 4, 401-414, 1988
Forster, A C, Jeffries, A C, Sheldon, C C, Symons, R H. Structural and ionic requirements for self-cleavage of virusoid RNAs and trans self-cleavage of viroid RNA. Cold Spring Harb. Symp. Quant. Biol., 52, 249-59, 1987
Forster, A C, Symons, R H. Self-cleavage of virusoid RNA is performed by the proposed 55-nucleotide active site. Cell, 50(1), 9-16, 1987
Forster, A C, Symons, R H. Self-cleavage of plus and minus RNAs of a virusoid and a structural model for the active sites. Cell, 49(2), 211-20, 1987
Symons, R H, Hutchins, C J, Forster, A C, Rathjen, P D, Keese, P, Visvader, J E. Self-cleavage of RNA in the replication of viroids and virusoids. J. Cell Sci. Suppl., 7, 303-18, 1987
Hutchins, C J, Rathjen, P D, Forster, A C, Symons, R H. Self-cleavage of plus and minus RNA transcripts of avocado sunblotch viroid. Nucleic Acids Res., 14(9), 3627-40, 1986
Forster, A C, McInnes, J L, Skingle, D C, Symons, R H. Non-radioactive hybridization probes prepared by the chemical labelling of DNA and RNA with a novel reagent, photobiotin. Nucleic Acids Res., 13(3), 745-61, 1985
Visvader, J E, Forster, A C, Symons, R H. Infectivity and in vitro mutagenesis of monomeric cDNA clones of citrus exocortis viroid indicates the site of processing of viroid precursors. Nucleic Acids Res., 13(16), 5843-56, 1985
Lab pics








UAG