Exp1: test for flipping and cross-talk
The first experiment is design to test if integrases and excisionases pairs can flip a target sequence in both directions in a controlled manner. Then, we will test if these integrases are specific by putting them in presence of non-target sites. If this first set of experiment is successfull, we will then perform a kinetic analysis of flipping, by varying time of induction and concentrations of recombinases.
In a first time we will characterize 3 Integrases/Excisionases pairs.
Experimental design
Steady state experiment: Does it flip ?
- organism
E.coli for simplicity
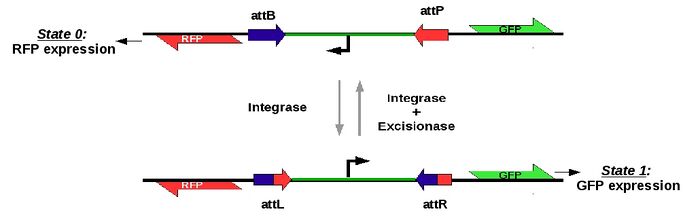
- Brief description
A target "flipper" plasmid bearing a T7 promoter flanked by specific attP and attB or attL and AttR recombination sites in antiparallel orientation will be cotranformed in e. coli cells with a plasmid expressing the corresponding integrase, or the integrase and excisionase pair. Each target sequence will be flanked on each side by a gene encoding a different fluorescent protein, enabling the monitoring of promoter orientation.
Cells will be allowed to grow and to reach steady stte of the reaction and the pourcentage of flipping cells will be determined.
In a second time, integrases and excisionases will be tested for their ability to act on non-cognate sites (cross talk).
- Constructs
First, we work with standards parts. We need to choose which Biobrick format we want to use (see here).
Standard chosen is Freiburg: allow protein fusion, compatible with BBa
Back to Integrase memory notebook