Endy:Notebook/DNA Sequence Refinement/Algorithm Proposal I
From OpenWetWare
Jump to navigationJump to search
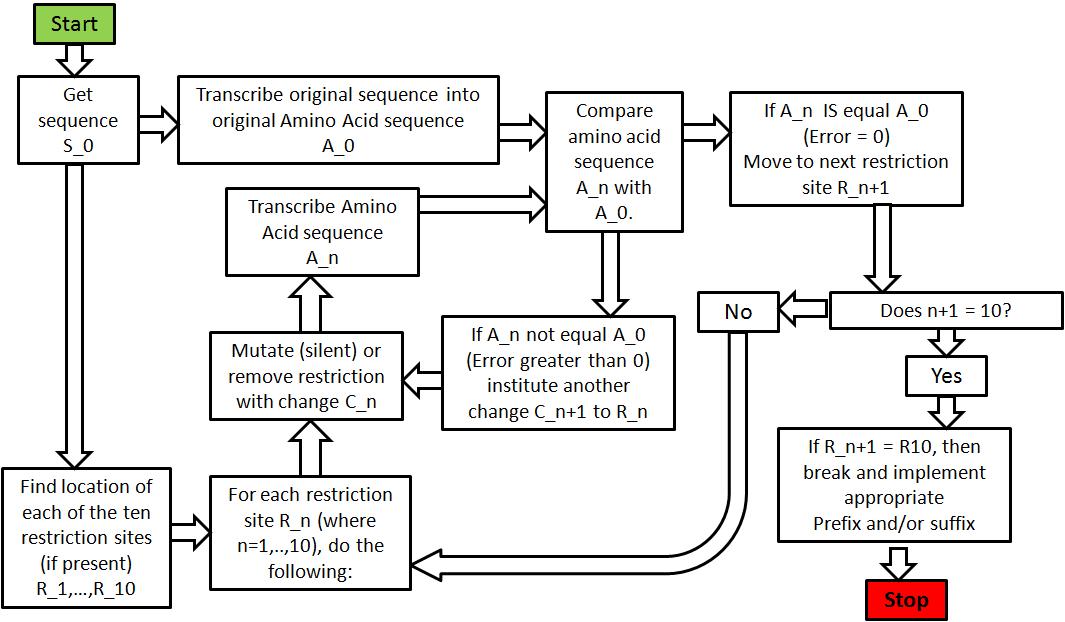
- Get desired original sequence S_0.
- Transcribe S_0 into an Amino acid sequence A_0.
- Search sequence S_0 to check if any of the ten specified restriction sites exist (EcoRI, XbaI, SpeI, PstI, NotI, PvuII, XhoI, AvrII, NheI and SapI).
- For each restriction site R_n, do the following:
a. Perform a change C_n (either silent mutation or deletion). b. Transcribe a new Amino acid sequence (A_n). c. Compare Amino acid sequence A_n with A_0.
i. If not the same, perform another change C_n+1 on the restriction site R_n and generate a new Amino acid sequence. Repeat until the generated Amino acid sequence is the same as A_0. ii. If the change C_n does not affect Amino acid transcription, move to the next restriction site R_n+1. d. Continue until all restriction sites had been modified and the Amino acid transcription has 1-to-1 parity with the original Amino acid sequence A_0. e. Add necessary prefixes/postfixes as stated by the appropriate BioBrick standard.