Endy:Measuring Screening Plasmid on MOFLO
From OpenWetWare
Jump to navigationJump to search
Setup the machine
- Set PMT voltages to FL1 = 525, FL7 = 650.
- This is the highest settings that eliminate "dark noise". Additionally, these setting are acceptable for the full range of arabinose induction for the current Screening Plasmid series. (I13534,I13537,I13538).
- If you are not sorting the droplet maker (technical name?), the stream can be turned off to reduce noise in the readings.
- The threshold for SSC should be set to zero (lots of noise) and then raised until you have a very low amount of noise. you can determine what is noise by cutting off the sample flow anything showing up then is machine noise. (the machine noise and the cells should be in very different positions on the FSC/SSC plot) The purpose of the low amount of noise is to make sure you're not cutting off any of the sample - e.g. you didn't go too high with the cutoff.
- You should only see a single population on the FSC/SSC plot.
- If you are getting very low counts it could be because the threshold is too high. Remember that you can tell the cells from the noise by closing the flow (yellow valve) and then seeing which population disappears.
- FSC can't be adjusted when set on log scale
- SSC 418
- The SSC knob is jumpy, pull on it slightly while you turn to get the number to stabilize.
- Pressure differential should be set so counts are less than 30,000 (this number is listed as 'trigger rate' (x1000) on the machine)
- The pressure differential (sample flow - sheath flow) can also effect the noise in the system, higher pressures have wider streams and so the cells may not be centered as well on the laser. You can measure the noise caused by the pressure differential by measureing the CV of measurements (FSC or FL1,2,etc) of standard beads (Flow Check by Beckmen Coulter). Glenn says ths CV for the MOFLO should be < 2%.
- Make sure the parameter scale for FL1/7 is set to log (on both the machine and the graphs on the computer).
Controls
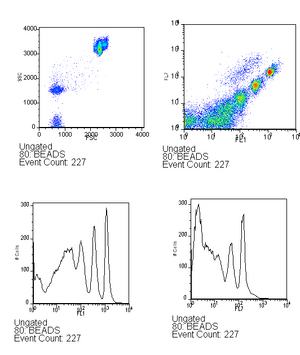
- Run beads (~6 drops in 500ml) - this is used to verify that the lasers are aligned as well as to serve as a calibration between runs, you can read more about this in the bead calibration page. Be sure to save the bead calibration data.
- Run negative control
- Run blank media
- Empty screening plasmid in high induction (to get a "high" GFP/RFP control)
- Experiment specific controls (often inlcudes the empty screening plasmid or construct expressiong GFP only, see specific protocol for details.)
Notes
Include links to control runs with M9, SuppM9, Negative control, etc. Include the eps (events per second) results as well.
Aquiring samples
- Back flush the collection tube (for at least 10 sec) to clear out the previous sample
- Open and close the yellow valve a few times in order to clear it out better.
- Vortex sample
- Load sample and collect cells (number depends on protocol)
- Close the red valve and yellow valve and wait for 5 seconfs before removing the tube, this lets the pressure go down and prevent the fluid from splashing in your face. (most of the time)
- Begin back flush immediately after removing sample (the longer it runs the better).
Notes
- If the cells begin arriving very slowly check that the laser power is still high. If it has declined karate chop the laser and it will fix it. (i'm serious).
Sorting samples
- Load cells as decribed above, you may want to run bleach followed by water (?) in between samples if you are concerned about contamination.
- Run the sample in "Aquire mode" first in order to save a file of the cell population (the files are not saved while sorting).
- When finished aquiring stop the cell from flowing by closing the yellow valve, then switch to sorting mode and define a sort region.
- Print the page with the sort region since as far as I know right now that's the only way to document it (there has to be a way, ask about it).
- Reset the sort count on the machine behind you
- Insert the collection tube with fresh media, be careful not to touch the plates! (technical name)
- Open the yellow valve
- Collect X cells (defined by specific protocol)
- Begin back flush immediately
Collecting Data
- Before burning data to a CD, check that none of the files have 0kb. There seems to be a bug in the Summit software so that occaisonally I lose and fcs file. If it's got 0kb just re-run the sample.
Questions
- Why does increasing the SSC threshold lead to counts in regions that aren't there when the threshold is set = 0? (e.g. as you increase the threshold you actually start to change the distribution).
- Is it because those counts are just there in very low frequency and the points on the screen are only some fraction of the total counts -- so when you threshold the bulk of them out you actually start to see things that would never have shown up at all?