Elizabeth Polidan Week8

Elizabeth Polidan
BIOL 398.03 / MATH 388
- Loyola Marymount University
- Los Angeles, CA, USA
5. Choose two genes from Figure 4.6 and draw a graph to represent the change in transcription over time.*Note: Dr. Dahlquist said that this will be done on a seperate piece of paper to be submitted in class on Thursday.
This will be submitted on Tuesday.
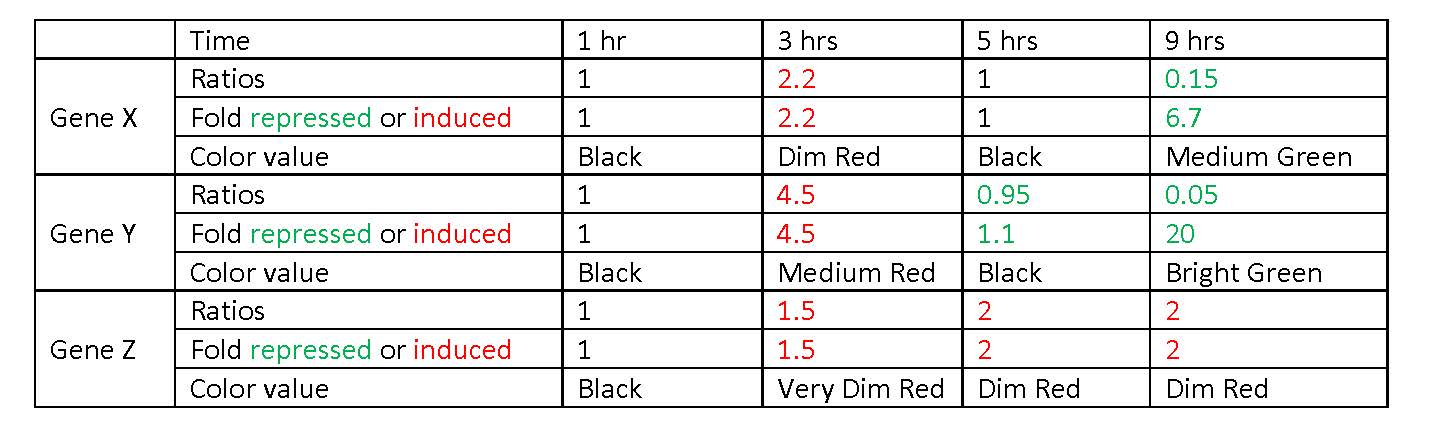
6b. Look at Figure 4.7, which depicts the loss of oxygen over time and the transcriptional response of three genes. These data are the ratios of transcription for genes X, Y, and Z during the depletion of oxygen. Using the color scale from Figure 4.6 (bright, medium, dim green, black, dim, medium, or bright red), determine the color for each ratio in Figure 4.7b.
This question falls on the pages before it discusses the log transform. Therefore I assume we are to use the ratio values and determine the fold repression of the values less than 1.0 (red) using the equation Fold repression = 1/ratio
7. Were any of the genes in Figure 4.7b transcribed similarly?
The closest similarity is that Genes X and Y start out repressed and then become induced between 3 and 5 hours. However, the curves of these ratios are not similar.
9. Why would most spots be yellow at the first time point?
At the first hour the oxygen level is still at 100% so there would be no environment pressure yet.
10. Go to http://www.yeastgenome.org and search for the gene TEF4; you will see it is involved in translation. Look at the time point labeled OD 3.7 in Figure 4.12, and find the TEF4 spot. Over the course of this experiment, was TEF4 induced or repressed? Hypothesize why TEF4’s gene regulation was part of the cell’s response to a reduction in available glucose (i.e., the only available food).
TEF4 is repressed over the entire course of the experiment (except for the first frame as noted in the caption). TEF4 regulates translation elongation factor activity, which means it is involved in the protein synthesis. When available food becomes scarce all unnecessary functions are shut down, so genes such as TEF4 are repressed.
11. Why would TCA cycle genes be induced if the glucose supply is running out?
The TCA cycle generates energy from sources other than glucose. Acetate derived from carbohydrates, fats and proteins is oxidized into carbon dioxide.
12. What mechanism could the genome use to ensure genes for enzymes in a common pathway are induced or repressed simultaneously?
One gene could regulate all of those genes when they are involved in that common pathway. They might be regulated by other genes for other functions.
13. Given rule one on page 109, what color would you see on a DNA chip when cells had their repressor gene TUP1 deleted?
You would see red because there was nothing to repress the gene expression.
14. What color spots would you expect to see on the chip when the transcription factor Yap1p is overexpressed?
You would see red because Yap1p regulates transcription activation, so you would see induced expression.
15. Could the loss of a repressor or the overexpression of a transcription factor result in the repression of a particular gene?
The only way I can see this happen is if there is another regulating gene and it represses the gene after it “over expresses.”
16. What types of control spots would you like to see in this type of experiment? How could you verify that you had truly deleted or overexpressed a particular gene?
Control spots should be genes that cannot be expressed except for in the presence of the deleted gene or other genes that need the overexpression of the gene in question.