Dahlquist:Lactase Persistence Genotyping by qPCR
This protocol is based off of Weinlander, K. M., Hall, D. J., & De Stasio, E. A. (2010). RFLP analysis and allelic discrimination with real‐time PCR using the human lactase persistence trait. Biochemistry and Molecular Biology Education, 38(3), 167-171. doi: 10.1002/bmb.20357 (full text)
Primers
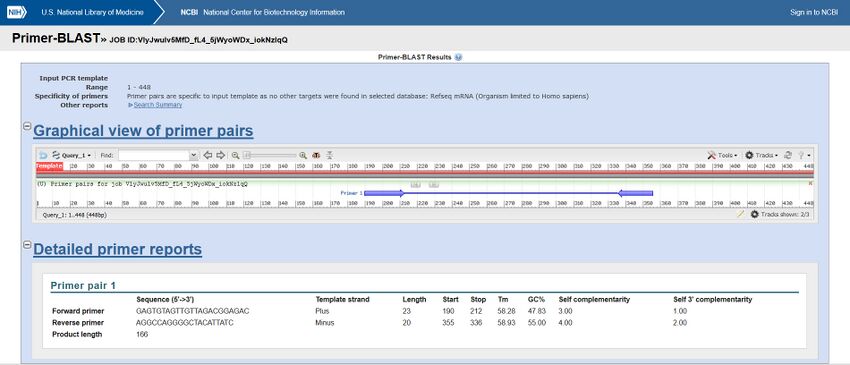
Forward primer sequence:
5'-GAGTGTAGTTGTTAGACGGAGAC-3'
qPCR primer sequence for T genotype:
5'-AGGCCAGGGACTACATTATC-3'
qPCR primer sequence for C genotype:
5'-AGGCCAGGGGCTACATTATC-3'
Reaction Mix
From Weinlander paper:
7 ng genomic DNA 1.4 µL of 3.5 µM forward primer 1.4 µL of 3.5 µM reverse primer C or T 0.5 µL 50X ROX 12.5 µL master mix Water to 25 µL
Note: the final concentration of each primer is 0.2 μM and the master mix contains modified Thermus brockianus DNA polymerase, dNTPs, SYBR Green, MgCl2, and buffer solution.
Modified from Apex qPCR GREEN Master Mix Manual:
Component Vol./reaction* Final concentration* Apex qPCR 2X Master Mix 12.5 μL 1X Primer A (10 μM) 0.5 μL (0.125 – 1.25 μL) 0.2 μM (0.05 – 0.5 μM)** Primer B (10 μM) 0.5 μL (0.125 – 1.25 μL) 0.2 μM (0.05 – 0.5 μM)** PCR-grade H2O 6.5 μL --- Template DNA 5 μL genomic DNA: 20 ng (1 – 100 ng) or plasmid DNA: 0.5 ng (0.1 – 1 ng) TOTAL volume 25 μL ---
* Suggested starting conditions; theoretically used conditions in brackets.
** Optimization of primer concentrations is highly recommended. Note that I had to change the final concentration to 0.2 μM from 0.1 μM because manual incorrect for the 0.5 μL volume. I changed the theoretical volumes of the primer to match the theoretical final concentrations.
Organizing Reactions
- Each sample (template) needs to be run with two different primer pairs, which means two master mixes will be made, one for each primer pair:
- Forward + Reverse (C)
- Forward + Reverse (T)
- Each sample + primer pair needs to be run in duplicate or triplicate as technical replicates to account for technical differences (pipetting error, etc.)
- There needs to be a negative no DNA template control (using water instead of DNA) for each primer pair/master mix
- We will use the plasmids as positive controls, "T" plasmid, "C" plasmid, and a mix of "T" and "C" plasmid. Each of these is run with both primer pairs.
- We should do a dilution series of 3-5 dilutions for each of these to determine the right concentration of plasmid template to use going forward.
Instrument Protocol
qPCR carried out in four stages run consecutively.
- Stage 1: 50.0°C for 2 minutes.
- Stage 2: 95.0°C for 10 minutes.
- Stage 3: 40 cycles of 95.0°C (for 15 seconds) and 60.0°C (for 1 minute).
Results for each sample were determined by selecting only the amplification plots of both tubes after the threshold was set.
Troubleshooting
- Sigma-Aldrich qPCR Guide
- Their recommended standard instrument protocol is:
- Initial denaturation 94°C for 2 min
- Denaturation 94°C for 15 sec
- Annealing, extension, and read fluorescence 60°C or 5°C below lowest primer TM for 1 min
- repeat steps 2 and 3 for 40 cycles
- has diagram regarding how SYBR works
- explains quantification cycle relative to amount of starting material (i.e. dilutions)
- lists supplies needed and components of assay and why each is important
- has troubleshooting guide
PrimerXL Design
This DMAS (double-mismatch allele-specific) assay was designed at PrimerXL.org, based on the paper:
- Lefever, S., Rihani, A., Van der Meulen, J., Pattyn, F., Van Maerken, T., Van Dorpe, J., ... & Vandesompele, J. (2019). Cost-effective and robust genotyping using double-mismatch allele-specific quantitative PCR. Scientific reports, 9(1), 2150. https://doi.org/10.1038/s41598-019-38581-z
Assay information
Ensembl build Release 75, build 37
Taxonomy ID 9606
Description rs4988235
Target rs4988235
Template gdna
Constant primer GCGAAGATGGGACGCTTGAA
Allele-specific primer 1 CGCTGGCAATACAGATAAGATAATGCAGC
Allele-specific primer 2 CGCTGGCAATACAGATAAGATAATGCAGT
Amplicon GCGAAGATGG GACGCTTGAA TGCCCTTTCG TACTACTCCC CTTTTACCTC
GTTAATACCC ACTGACCTAT CCTCGTGGAA TGCAGGGCTC AAAGAACAAT
CTAAAAATCA AACATTATAC AAATGCAACC TAAGGAGGAG AGTTCCTTTG
AGGCCAGGGG CTACATTATC TTATCTGTAT TGCCAGCG
Length 188
Amplicon location 2:136608487-136608674
Design settings SNP analysis relaxed Sec. struct. analysis relaxed Specificity analysis yes Primer annealing Tm (min-opt-max) 63.00-65.00-67.00 Primer GC% (min-opt-max) 20.00-50.00-80.00 Amplicon length range 80-250 # G/C's in last 5 3' primer bp 5 Mg concentration 0.05 Na concentration 0.003 DNA concentration 250 dNTP concentration 1.2 PCR reaction temperature 60
Double-stranded 188 bp amplicon (primers bolded) 5’-GCGAAGATGG GACGCTTGAA TGCCCTTTCG TACTACTCCC CTTTTACCTC GTTAATACCC ACTGACCTAT CCTCGTGGAA TGCAGGGCTC-3’ 3’-CGCTTCTACC CTGCGAACTT ACGGGAAAGC ATGATGAGGG GAAAATGGAG CAATTATGGG TGACTGGATA GGAGCACCTT ACGTCCCGAG-5’ 5’-AAAGAACAAT CTAAAAATCA AACATTATAC AAATGCAACC TAAGGAGGAG AGTTCCTTTG AGGCCAGGGG CTACATTATC TTATCTGTAT-3’ 3’-TTTCTTGTTA GATTTTTAGT TTGTAATATG TTTACGTTGG ATTCCTCCTC TCAAGGAAAC TCCGGTCCCC GATGTAATAG AATAGACATA-5’ 5’-TGCCAGCG-3’ 3’-ACGGTCGC-5’
- "Blue" nucleotide is the site of the SNP (C/T).
- "Red" nucleotide indicates DMAS mismatch in the 4th position from the 3' end of the allele-specific primer.
Relationship to 448 bp fragment from RFLP assay
448 bp RFLP fragment (sequence in DMAS amplicon red) GGATGCACTG CTGTGATGAG GTATCAGAGT CACTTTGATA TGATGAGAGC AGAGATAAAC AGATTTGTTG CATGTTTTTA ATCTTTGGTA TGGGACATAC TAGAATTCAC TGCAAATACA TTTTTATGTA ACTGTTGAAT GCTCATACGA CCATGGAATT CTTCCCTTTA AAGAGCTTGG TAAGCATTTG AGTGTAGTTG TTAGACGGAG ACGATCACGT CATAGTTTAT AGAGTGCATA AAGACGTAAG TTACCATTTA ATACCTTTCA TTCAGGAAAA ATGTACTTAG ACCCTACAAT GTACTAGTAG GCCTCTGCGC TGGCAATACA GATAAGATAA TGTAGCCCCT GGCCTCAAAG GAACTCTCCT CCTTAGGTTG CATTTGTATA ATGTTTGATT TTTAGATTGT TCTTTGAGCC CTGCATTCCA CGAGGATAGG TCAGTGGG Reverse complement of 188 bp DMAS amplicon (sequence in RFLP fragment red) CGCTGGCAAT ACAGATAAGA TAATGTAGCC CCTGGCCTCA AAGGAACTCT CCTCCTTAGG TTGCATTTGT ATAATGTTTG ATTTTTAGAT TGTTCTTTGA GCCCTGCATT CCACGAGGAT AGGTCAGTGG GTATTAACGA GGTAAAAGGG GAGTAGTACG AAAGGGCATT CAAGCGTCCC ATCTTCGC
BLAST results
Score Expect Identities Gaps Strand
243 bits(131) 5e-69 131/131(100%) 0/131(0%) Plus/Minus
Query 318 CGCTGGCAATACAGATAAGATAATGTAGCCCCTGGCCTCAAAGGAACTCTCCTCCTTAGG 377
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 188 CGCTGGCAATACAGATAAGATAATGTAGCCCCTGGCCTCAAAGGAACTCTCCTCCTTAGG 129
Query 378 TTGCATTTGTATAATGTTTGATTTTTAGATTGTTCTTTGAGCCCTGCATTCCACGAGGAT 437
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 128 TTGCATTTGTATAATGTTTGATTTTTAGATTGTTCTTTGAGCCCTGCATTCCACGAGGAT 69
Query 438 AGGTCAGTGGG 448
|||||||||||
Sbjct 68 AGGTCAGTGGG 58