Constructing the Plasmids
Fusion PCR
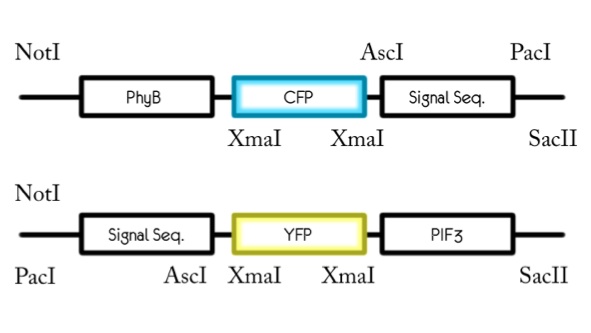
Construction of the PhyB-CFP and Pif3-YFP Plasmids began with the amplification of each individual sequences, PhyB, CFP, Pif3 and YFP, from the Quail Lab Plasmids with PhyBNT-GalBD and GalAD-PIF3. Through Fusion PCR the PhyB-CFP and Pif3-YFP sequences were then created. These sequence were subsequently inserted into the prS313 backbone using the Not1, SacII enzymes, a plasmid which contains a constitutive Myo2 promoter and the His gene.
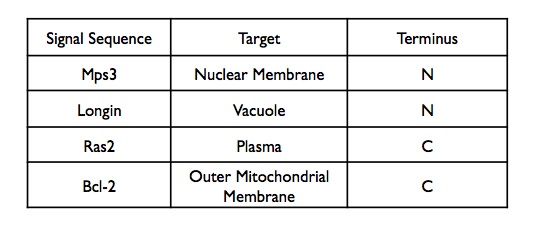
A variety of C and N terminus localization Sequences were chosen to test the PhyB, Pif System. Each localization sequence, once amplified, was cloned into the appropriate backbone using the AscI, PacI enzymes. N terminus tags were destined for the Pif3-YPF backbone and C terminus for the PhyB-CFP Backbone.
Sequencing the PhyB-CFP constructs and YFP-PIF3 constructs failed. Our sequence did not match what we expected.
Homologous Recombination
Forward and reverse primers were designed for amplification of PhyB, CFP, YFP, and PIF3. The primers were designed so that 40bp regions of homology of both the backbone and the other construct were attached to the ends
PhyBNT hr primers: Sense: 40bp homology with pRS313, Antisense: 40bp homology with CFP
CFP hr primers: Sense: 40bp homology with PhyBNT Antisense: 40bp homology with pRS313 and AscI,PacI sites for cloning signal sequences.
YFP hr primers: Sense: 40bp homology with pRS415 and AscI, PacI sites for cloning signal sequences. Antisense: 40bp homology with PIF3
PIF3 hr primers: Sense: 40bp homology with YFP Antisense: 40bp homology with pRS415
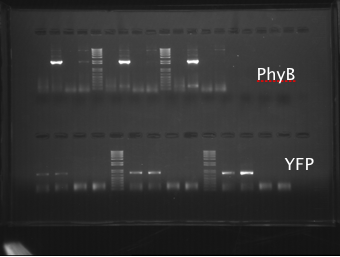
PhyBNT, CFP, YFP and PIF3 were PCR amplified, and the PCR products were analyzed on the gel shown below.
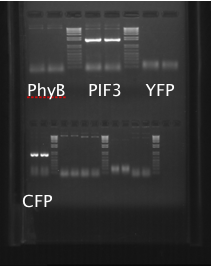
PIF3 and CFP successfully amplified, but PhyB and YFP did not. PIF3 and CFP PCR products were gel purified (gel shown below).
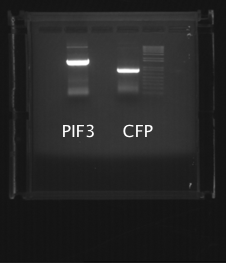
PCR was then attemped again, at various primer concentrations and at different temperatures. The PCR products were analyzed using gel electrophoresis (shown below), showing that PhyBNT and YFP successfully amplified.
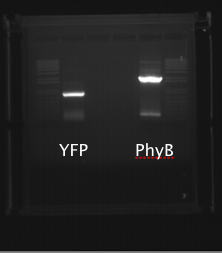
PhyBNT and YFP were gel purified (gel shown below).
pRS313 with Myo2 promoter and pRS415 with ADH1 promoter were digested with NotI and SacII. The linearized DNA was gel analyzed and purified (gel shown below).

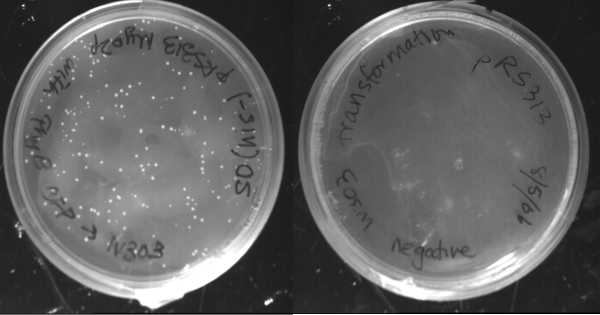
The digested pRS313 Myo2p was transformed into yeast along with the purified PhyBNT and CFP products. As a negative control, the digested pRS313 Myo2p was transformed without the PhyBNT and CFP. The transformed cells were plated onto his- selective plates. We see many more colonies on the plate with the PhyBNT and with the CFP inserts than without, suggesting that the inserts have been added into the plasmid via homologous recombination.
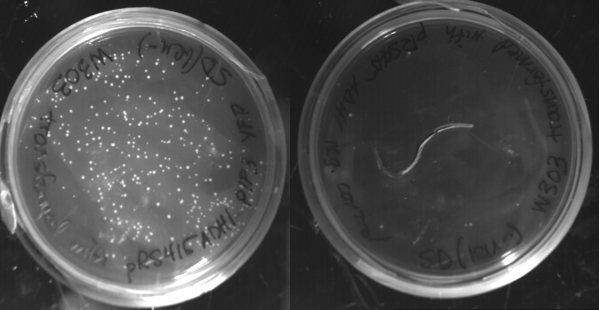
A similar procedure was conducted with the YFP and PIF3 inserts and the pRS415 ADH1 backbone. The digested pRS415 ADHI was transformed with the YFP and PIF3 products. As a negative control the digested pRS415 ADH1 was transformed without the YFP and PIF3 products. The transformed cells were plated onto leu- selective plates. The presence of more colonies on the plate in which the inserts were added than on the one where the inserts weren't added show that the inserts have been cloned into the plasmid via homologous recombination.
Colonies from the plates with the inserts were minipreped using a modified QIAGEN protocol for minipreps in yeast. The plasmid were then electroborated into E. coli and bacterial minipreped. The DNA product was sequenced, showing that the expected constructs were built, however with a few point mutations in the sequence. Liquid culture from the yeast colonies that had been picked were analyzed using fluorescent microscopy. No fluorescence was observed.