CH391L/S13/Optogenetics
Introduction to Optogenetics
"Before we can find the answers, we need the power to ask new questions." -Karl Deisseroth
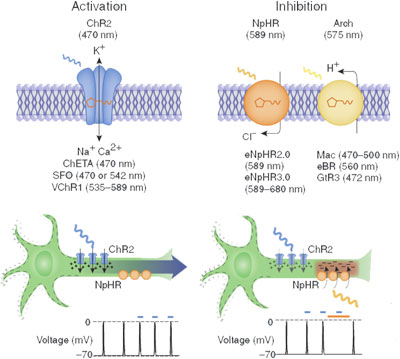
The umbrage of optogenetics encompasses a plethora of techniques in which light sensitive membrane proteins are transferred into nonnative systems in order to elicit a biochemical event such as transcription, kinase activity or even ion transfer. Today, the term "optogenetics" tends to be associated with the field of neuroscience, where arguably optogenetics proves to be the most utilitarian. The wide array of opsins and photoreceptors delineate into a distinctive set of proteins with special characteristics. These proteins provide a viable an inducible transduction system for scientist to take advantage of already natural biochemical pathways.
In brief, the neuron in question is spliced with a specific opsin gene carried by viral vector, usually a modified lentivirus. Subsequently, the encoded opsin attaches to the cell membrane. Opsin are photosensitive G protein receptors or ion channels that are induced by a specific wavelength of visible light via fiber optic cable or optrode. In turn, the opsin undergoes a conformational change, eliciting a change in membrane potential. For neuronal cells, changes in membrane potential give rise to action potentials. The strength of the depolarizing current (incoming positive charged ions) is encoded in the frequency of the action potentials generated. Furthermore, synapses (gap junction between neurons) can conduct spatial or temporal summation. Even neuronal cells can be silenced with hyperpolarizing current (incoming negative or outgoing positive charged ions). In short, optogenetics provides neuroscientists with an “on/off” switch for targeted neurons.
In essence, optogenetics is a neural modulation technique used to control neurons in vitro for the purpose of affecting the physiology of neural circuits and ultimately behavior of the studied organism. However, in recent studies, optogenetic techniques have been used to modify nonneuronal tissues, such as cardiac tissue and beta cells, willfully controlling the respective cell-specific roles. Although the implications of optogenetics seem like a panacea for many genetic diseases, much of the field is new; in terms of therapeutics, research is regretfully far behind. As of now, optogenetic techniques are attempted mostly on rodent specimens since primate studies lack profound electrophysical and behavior effects. Current optogenetically-related research aims to ascertain brain function of multiple neural circuits, but future endeavors include gene therapy for neurodegenerative diseases and neuroprosthetics.
History
"The brain is a world consisting of a number of unexplored continents and great stretches of unknown territory." - Santiago Ramón y Cajal
| Year | Event |
|---|---|
| 1971 | Bacteriorhodopsin (yellow and green) found in Halobacterium halobium |
| 1977 | Halorhodopsin (yellow) found in Halobacterium halobium |
| 1999 | Francis Crick's Declaration |
| 2002 | ChR1 (blue) discovered in Chlamydomonas reinhardtii, Zemelman and Miesenbock chARGed optogenetic system [2] |
| 2003 | ChR2 (blue) discovered in Chlamydomonas reinhardtii |
| 2005 | Millisecond-timescale, genetically targeted optical control of neural activity [3] |
| 2006 | "Optogenetics" coined |
| 2008 | VChR1 found in Volvox carteri (yellow and possibly green) |
The story behind optogenetics is a story of convergence of two seemingly unrelated fields: microbial microbiology and neuroscience. In 1971, Walther Stoeckenius and Dieter Oesterhelt discovered Halobacterium halobium contained transmembrane protein bacteriorhodopsin (BR) that is capable of moving hydrogen ions into the cell due to the presence of light. BR is highly expressed in haloarcheal membranes while a similar transmembrane protein proteorhodopsin (PR) is expressed in eubacteria.
In 1977, Matsuno-Yagi and Mukohata described the ion pump channel halorhodopsin which displaces extracellular chloride ions in to the cell. As these discoveries were happening in the background, a growing desire to effectively control the neurons was ever prevalent. During this period, neuroscientists adopted the idea that cognitive processes were distributed in various localizations of the cortex. This fact was evident when lesions in specific areas of the brain would lead to loss of a specific process.
fMRIs and PET solidified this point, but even with these high-tech instruments, the processes themselves could not fully be understood. This would require an in-detail analysis of the neural circuits that make up the localized cortical space. This would entail the use of electrodes in order to stimulate specific cells to generate action potentials. Electrode analysis and other biochemical methods provided much of the insight that we currently have on neuroscience, but this systematic approach is inherently flawed. Even though the electrodes are made extremely small to excite a particular cell, the electric charge permeates the cerebral space so that other close neurons are also stimulated. Such a problem defeats the purpose of ceteris paribus, since we are unable to determine whether the cell in question elicited the desired response in the first place. Moreover, to conduct a loss of function experiment, the neural tissue is question needs to be destroyed, or lesioned, but, once the tissue is destroyed, there are no methods to restored said tissue, further complicating neural circuit research.
This lack of cell specificity and cellular controls encouraged neuroscientists to find alternative methods. In the 1990s, some scientists became infatuated with the idea that opsins may be the key to cell specificity. However, for the most part, this idea was considered too “far-fetched” to allocate resources, time, and money.
In 1999 at the University of California in San Diego, Francis Crick commented that neuroscience faced a major challenge: controlling one type of cell while leaving the others unaffected. Crick even suggested that light could serve as a control tool. In 2002, Boris Zemelman, currently a UT associate professor, and Gero Miesenbock conducted the earliest genetically targeted method with rhodopsin photoreceptors in Drosophila[2].
Also in 2002, channelrhodopsin (ChR1) was identified by Georg Nagel to contain a wider cation conductance (Na and K ions).
In 2003, Channelrhodopsin-2 (ChR2) was later discovered in the same organism Chlamydomonas reinhardtii which conducts cations faster than ChR1 (twofold greater rate). ChR2 became the key to excitatory neuron transmission. In 2005, Karl Deisseroth, Edward S. Boyden, and Feng Zhang created a robust ChR2 optogenetic system cultured in mammalian neurons for precisely timed light driven action potentials[3]. The term “optogenetics” was coined in 2006[1].
Biochemistry behind Optogenetics
The appeal of optogenetics stems from the simplicity of opsin; the opsin proteins combine the two roles of light sensitivity and ion transport while only being encoded by a single gene. Optogenetics takes advantage of opsins’ ability to activate for various wavelengths of light and the ubiquitous presence of retinal in vertebrate tissue, including the brain. All types of opsin require retinal as a cofactor that serves as a antenna for photons. When the cofactor retinal is bound, the opsin protein is termed rhodopsin. After the retinal has diffused into the binding pocket, it is covalently attached to a lysine residue on helix 7 by the formation of protonated retinal Schiff base (RSBH+). The ionic environment of the RSBH+ determines the spectral and kinetic properties of the particular opsin. Indeed, this very characteristic has been exploited to create mutant opsin proteins in vitro through mutagenesis analysis. Also, all opsins consist of a seven-transmembrane helix oval. Furthermore, opsin proteins are typically divided into two distinct families: type I and type II. Type I consists of opsin proteins found in prokaryotes, algae, and fungi, while Type II consists of opsin proteins found in higher eukaryotic organism such as those found in the human eye. Type I begins with all-trans configuration of retinal then isomerizes to 13-cis configurations, while type II isomerizes 11-cis retinal into all-trans retinal. Unlike type II, type I opsins do not dissociate from the retinal cofactor.
As previously discussed, bacteriorhodopsin (BR) was the first discovered opsin. Once light hits BR, the protein undergoes a series of protein confirmations between states that absorb light at 568 nm and 400 nm. Also, the moment light strikes the opsin to the release of the hydrogen ion to the extracellular environment is known as a complete photocycle. Each photocycle is made up of multiple photointermediates with different spectral characteristics and represents a separate proton translocation process.
BR process:
- Photon absorption initiates isomerization of bound retinal from all-trans to 13-cis
- Confirmational shift and dipole of RSHB+ raises pK, releasing RSB proton (deprotonated retinal Schiff base) and created M intermediate (410nm maximum absorbance)
- Release RSB proton is accepted by the Asp 85
- Simultaneously, a different proton is released from an amino acid group from the proton release complex (PRC)
- RSB captures proton from Asp 96 and the protein enters the N intermediate (560 nm)
- Asp 96 is repronated by cytoplasmic protons while the Schiff bases reisomerizes into all-trans retinal (630nm)
- Asp 85 transfers its proton to the PRC[4]
In the case of halorhodopsin (HR), the photocycle does not include the depronation of RSBH+. After light induction, the proton cannot be releases because the acceptor amino acid normally found is BR has been replaced by Threonine. Instead, a chloride ion is moved from through the chromophore and then sifted through the protein. In perspective of neuroscience, the expression of inhibition requires additional mammalian membrane trafficking signals, because the inward current provided by HR activation is relatively weaker than the normal depolarization seen in most neural cells. Also, HR is activated by yellow light[4].
Sensory rhodopsin (SR-I) and phoborhodopsin (SR-II) are similar to BR except the opsin activates secondary messager Htr to activate an associated phosphorylation cascade.
Channelrhodopsin (ChR) is also similar in homology with BR. ChR has three distinct forms: ChR1, ChR2 and VChR1. SFO mutation at Cys 128 in ChR2 leads to a twofold conductance of cations. A histine to arginine mutation in the proton donor (similar to Asp 96) leads to increased photocurrents but slows deactivation in ChR2. Chimeric proteins of ChR1 and ChR2 reduce inactivation and vary on photocurrent activity. Modifications on the proton acceptor (similar to Asp 85) in Glu 123 lead to more rapid channel closing and fast optical control of action potentials, while inactivation time is reduce- ChETA mutation. VChR1 absorbs light at a red-shifted maximum of 540nm while ChR1, ChR2, and VChR2 absorb blue light[4].
The Optogenetic Process
- Opsin gene is combined with a promotor to ensure the gene is active only in a specific type of cell
- Modified gene is inserted in the host brain via viral vector
- The virus infects many nerve cells but promoter only allows selected neuron to make opsin protein
- Fiber optic or optrode is inserted into brain and flashed the specific wavelength of light
- Record transformed regions
Gene Delivery
Opsin genes are transported into the target cells by transfection, viral transduction or transgenic animal lines. Although the viral delivery system isn’t cell type specific, the included promoter ensures expression only in the desired cells. In other words, the virus infects plethora of cell types, but the desired cells express the distinct promoter regions not shared by other neurons. Both lentivirus and adeno-associated virus vectors are used to transfer optogenetic circuitry into the desired cell type. Basically, the virus is loaded up with the promoter associated with the desired neuron and injected via stereotaxic needle closest to the desired region. Both lentivirus and AAV vectors induce long-term expression. However, lentivirus delivery are permanently integrated into the genome, while the AAV vectors are maintained extra-chromosomally. Even though virus delivery systems provide cell specific selection, the virus, themselves, have a limited packaging capacity. The lenitivirus can accomadate up to 10 kb while the AAV can include up to 5 kb. In response, transgenic animals can allow for bigger promoters to be used for increased specificity and increased expression[6].
Controlled Illumination
The basis of optogenetics is to use light to elicit a physiological response within the cell. This entails the uses of technologies that can illuminate transformed regions. In brief, these technologies include, LED, one-photon laser, optical fibers, and optrodes[6].
Record
After inducing the neurons with light, scientist must record the apparent behavior of the neural circuits by using electrodes, voltage sensitive fluorescent protein, and behavioral assays.
Applications of Optogenetics
Neuroscience
Obviously, optogenetics has huge applications in the field of neuroscience. Indeed, the ability excite and silence cells in order to ascertain the functional units of studied neural circuits are unparalleled, but the ultimate goal is to construct robust therapeutic technologies. For the example, parvalbumin, dopamingernic, and orexinergic neurons are affected by schizophrenia, Parkinson's disease, and narcolepsy, respectively. Deep brain stimulation is ineffective for such diseases, because the true onset of each disease is still unknown; random stimulation proves to be futile. In this respect, optogenetic research aims to further our understanding of neural diseases[7].
For example, epilepsy is a brain disorder in which the person has multiple seizures due to hyperactivity of certain cortical regions. Treatment for epilepsy is quite limited; current drugs and therapies do not properly treat or prevent the convulsions. Now, optogenetic research aims to target those hyperactive neurons and insert eNpHR3.0 opsin gene into the cells. In turn, the shining of yellow light would perturb the hyperactive neurons, effectively silencing them during an epileptic attack. This technology is analogous to an inhaler "silencing" an asthma attack[8].
Chimeric proteins, known as OptoXRs, composed of bovine rhodopsin and the G protein–coupled receptors allow optical control of biochemical mediated cascades.
Recently, by using optogentic tools, scientist have restored vision to blinded mice, by splicing ChR2 protein into the retina of the mice. Through rAAV, the opsin genes were transformed into ON bipolar cells[9].
IGEM Take-home Message
The 2012 IGEM team from the University of Washington conducted a project under optogenetics. The team's main focus was to effectively characterize the light sensor gene systems: Ccas and lavTAP. Both gene systems were combined with a lacZ system in order to determine the activity of the light sensor's autophosphorylation process. For the Ccas, green light stimulated autophosphorylation and expression of lacZ, while red light stimulates a conformational change that resists autophosphorylation and thus don't all binding to the promoter of the the lacZ. The lavTAP system is combined with an inverter gene so that blue light stimulates expression of the lacZ, analogous to repression regulation. In the future, the use of light sensors would benefit synthetic biologists by creating light sensor/ protein fusions to mediated specific biochemical cascades. [1]
Other Light Sensors
Light responsive proteins or "light sensors" are currently used as an method of signal transduction in order to regulate genetic circuits non-invasively within organisms. Light sensor proteins are found in all domains of life and have been utilized in complex genetic regulation to willfully turn off or on a gene of interest. Incidentally, the light sensor protein is a transmembrane channel or receptor found directly on the surface of most organisms. Particularly, the the majority of these light sensors consist of photoreceptors. In brief, a photon hits the photoreceptor and initiates a conformational change leading to possible phosphorylation by kinase or delocalization of trascription factor. Subsequently, this creates a biochemical signalling cascade that induces a molecular mediatory molecule to interact with the following regulatory molecules or simply interact with the gene complex. Photoreceptors are multidomain effector proteins that contain the protein component, pigment, and the chromophore. The pigment acts as a antennae station to attract photons. Photon acquirement lead to a change in the aromatic structure of chromophore which initiates the comforational change. By creating chimeras and other fusion proteins, an individual can easily take advantage of an promoter/ regulatory molecule to build new cascades thereby affecting different genetic ciruits[10].
Types of Photoreceptors
- rhodopsins- retinal chromophore, visual light used in eye cells.
- phytochromes- bilin chromophore, red light detection used by plants for circadian rhythm.
- xanthopsins- trans-p-coumaric acid chromophor, blue light avoidance response used by Ec. halophila
- cryptochromes- cryptochrome chromophore, blue-green light used by animals for circadian clock to seed germination in plants
- phototropins- Flavin chromophore (FMN), blue light for signal transduction in algae and bacteria
- BLUF- Flavin chromophore (FAD), blue light signal transduction in proteo- and cyano-bacteria [11]
References
- Zemelman BV, Lee GA, Ng M, and Miesenböck G. Selective photostimulation of genetically chARGed neurons. Neuron. 2002 Jan 3;33(1):15-22. DOI:10.1016/s0896-6273(01)00574-8 |
- Boyden ES, Zhang F, Bamberg E, Nagel G, and Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci. 2005 Sep;8(9):1263-8. DOI:10.1038/nn1525 |
- Yizhar O, Fenno L, Zhang F, Hegemann P, and Diesseroth K. Microbial opsins: a family of single-component tools for optical control of neural activity. Cold Spring Harb Protoc. 2011 Mar 1;2011(3):top102. DOI:10.1101/pdb.top102 |
- Mei Y and Zhang F. Molecular tools and approaches for optogenetics. Biol Psychiatry. 2012 Jun 15;71(12):1033-8. DOI:10.1016/j.biopsych.2012.02.019 |
- LaLumiere RT. A new technique for controlling the brain: optogenetics and its potential for use in research and the clinic. Brain Stimul. 2011 Jan;4(1):1-6. DOI:10.1016/j.brs.2010.09.009 |
- Kokaia M, Andersson M, and Ledri M. An optogenetic approach in epilepsy. Neuropharmacology. 2013 Jun;69:89-95. DOI:10.1016/j.neuropharm.2012.05.049 |
- Doroudchi MM, Greenberg KP, Liu J, Silka KA, Boyden ES, Lockridge JA, Arman AC, Janani R, Boye SE, Boye SL, Gordon GM, Matteo BC, Sampath AP, Hauswirth WW, and Horsager A. Virally delivered channelrhodopsin-2 safely and effectively restores visual function in multiple mouse models of blindness. Mol Ther. 2011 Jul;19(7):1220-9. DOI:10.1038/mt.2011.69 |
- Levskaya A, Chevalier AA, Tabor JJ, Simpson ZB, Lavery LA, Levy M, Davidson EA, Scouras A, Ellington AD, Marcotte EM, and Voigt CA. Synthetic biology: engineering Escherichia coli to see light. Nature. 2005 Nov 24;438(7067):441-2. DOI:10.1038/nature04405 |
- van der Horst MA and Hellingwerf KJ. Photoreceptor proteins, "star actors of modern times": a review of the functional dynamics in the structure of representative members of six different photoreceptor families. Acc Chem Res. 2004 Jan;37(1):13-20. DOI:10.1021/ar020219d |