BjornsMethods/DevelopmentPrinciples
3D Spatial Programming
Problem: How to program 3D spatial structures:
- Is it possible to discover and utilize the proper hierarchy of organization and dynamics to compile global 3D spatial structures through the use of smaller cellular objects? --> Biology does it. Can we learn from developmental biology?
Sub-Problems:
- What are the levels of abstraction?
- What are the interfaces between the levels of abstraction?
Definitions:
- Agent = individual cell
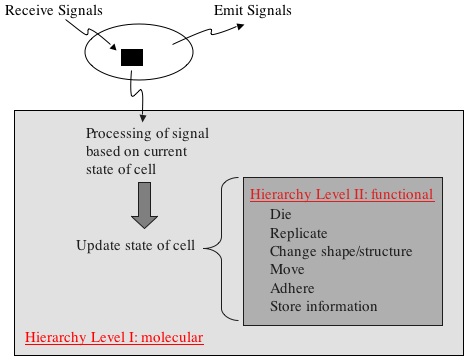
Hierarchy Level II: Agent Behavioral Primitives
- replicate (cell division)
- die
- change shape/structure
- crawl
- adhesion to substrates/other cells
- emit chemical signals (diffusible or membrane bound)
- receive chemical signals via receptors; also electrical and mechanical sensors?
- store information (memory)
- process signal and current state information
- NOTE: All of the above primitives are accomplished via a lower level of abstraction: biochemical primitives
Hierarchy Level I: Biochemical Primitives
- Gene Regulation
- Transcription Factors
- activators
- inhibitors
- Chromatin modification ==> global gene accessibility
- Methylation
- Acytlation
- Transcription Factors
- Protein modifications producing allosteric structural changes influencing protein function (Ex: on/off switch, change functionality)
- Kinases/Phosphatase (Phosphorylation state)
- Nucleotide Exchange Factors (GEF's, ATP additions, etc...)
- Methylation & Acytlation
- Polymerization of identical subunits (Ex: tubulin --> Microtubules)
- Dimerization (homo/hetero-)
- Protein complexes to perform higher level function not capable via single proteins
Hierarchy Diagram
Differentiation
One salient difference between amorphous computing and bio-cellular computing is that biology has devised an elegant hierarchical differentiation scheme to allow specialization and sub-specialization of cell types to distribute functions to a non-homogenous group of agents. Ultimately we want to meld differentiation into an amorphous-like computing system to utilize the power of hierarchical organization an heterogeneous populations.
A key component in the process of differentiation in biology is the idea I call a decision network. The purpose of a decision network is to perform a computation between communicating cells to determine the fates of the cells involved. A simple example of a decision network is the Delta-Notch ligand/receptor system found in nearly all organisms that performs a lateral inhibition role to prevent all cells within a local environment from differentiating into the same cell type. It is essentially a competition network where one cell wins and then subsequently inhibits the losers. Another decision network example is found in Dictyostelium prespore/prestalk (Psp/Pst) cell differentiation which is composed of a small negative feedback network acting as a homeostat that robustly produces a particular ratio of Psp/Pst cells (~75/25) within a given colony.
The utilization and understanding of decision networks are going to be a key interface in ultimately programming cellular differentiation within colonies of similar cells.
I argue that once a cell has made a decision what fate to take on, it wastes no time to act on that decision. For example, it is generally excepted that once a cell has decided it will commit suicide it will enter a fixed-action pattern (term stolen from behavioral neuroscience) to follow through to completion. What good is a half dead cell? Furthermore in Dictyostelium, once the cells have decided they will enter the social phase of their life, they immediately switch over to a new gene expression state (~25% of gene expression is modulated). This gene expression modulation occurs many times as the cells proceed with different morphogenetic transformations through development, but maintain a relatively stable gene expression pattern while no morphogenetic transformations are occurring. If it is true that significant state transitions occur in a crisp, decisive manner then it gives hope that genetic programming may be possible through the interaction of small decision networks and more global gene expression modules.
If you haven't noticed yet, I have absolutely no idea what I'm talking about and encourage any input/comments via email to: millard@mit.edu
Some Principles of Developmental Biology
Original List:
- Life vs. Death
- Cell proliferation (replication)
- Stem Cells - pleuri-potent cell replacers
- Cell death (apotosis)
- NOTE: Need both proliferation and death
- Cell proliferation (replication)
- Differentiation
- Cells change function and structure in a hierarchical way to build up a complex organism originating from a single cell
- Cell morphogenesis
- Cell shape/structure change to facilitate global morphogenesis
- Hierarchical Organization
- Cell differentiation results in reduction of cell potential
- Many possible cell fates all stemming from initial zygote (root of tree)
- Induction
- Cell communication that alters/induces cell fates
- Often times there are inducing centers --> a particular cell or group of cells emitting the induction factor(s)
- Community effect - if cell placed in certain environment, the cell may be induced to take on same fate as its neighbors
- Semi-modularity
- Sub-division of tasks (tissues and organs), help organize functional units
- Boundary formation - creates compartments to further order cells
- Cell sorting
- Cells can sort into groups of like cells through selective adherence to each other and to the extracellular matrix
- Strong Attractor Systems
- Allow fate determination to be relatively stable once arrived
- Competency
- Differential cell abilities to respond to signals due to its current state
- Epigenetics
- Chromatin structure and regulation of gene expression --> mechanism for changing competency and differentiation
- Combinatorial code of gene expression
- With limited number of genes/signals cells reuse same signals to mean different things given different competencies of cells listening
- Maternal factors
- Inherited directly from mother directly from mother cell
- Inherited regulatory molecules can effect cell fate autonomously without induction (determinants)
- Lateral inhibition
- Neighbors prevented from taking on same fate
- Analog to digital conversion through nonlinear thresholding
- Cells respond differently to the same signal when it is presented at different concentrations (morphogen).
- Continuous information contained within a concentration gradient is converted to a discrete outcome through thresholds of activation.
- Cell migration (motility)
- Often regulated by signaling molecules
- Cell adhesion
- Cells capable of adhering/pulling on other cells, surfaces, and extracellular matrices through surface proteins
- Cell sensing of self and environment
- Senses and interprets chemical, electrical and mechanical signals from other cells and environment
- Cellular information processing
- Cells process information based self state and environmental state
- Morphogenesis
- Taking on 3D form
- Typically embryos form 3D structures by first forming 2D sheets and subsequently folding
- 3D structure is often required for organ functionality
- Genetic pattern formation
- Spatially and temporally regulated differential gene expression
- Stimulatory vs. Inhibitory signaling (positive & neg feedback)
- Evolution
- Conservation of certain gene functions through evolution (homologs, orthologs)
- Cell memory
- Cell has ways of remembering past events by changing its current state based on experience