BioSysBio:abstracts/2007/Vincent Rouilly
Registry of BioBricks Models using CellML
Author(s): Vincent Rouilly1, Barry Canton2, Poul Nielsen3, Richard Kitney1
Affiliations: 1Imperial College London, 2MIT, 3The University of Auckland
Contact:email: vincent(dot)rouilly(at)ic(dot)ac(dot)uk
Keywords: 'synthetic biology' 'biobrick' 'cellML' 'model database'
Introduction
One of the main goals in Synthetic Biology is to assess the feasibility of building novel biological systems from interchangeable and standardized parts. In order to collect and share parts, a Registry of standardized DNA BioBricks has been established at the MIT. BioBricks can be assembled to form devices and systems to operate in living cells.
Design of reliable devices and systems would benefit from accurate models of system function. To predict the function of systems built from many parts, we need to have accurate models for the parts and mechanisms to easily compose those part models into a system model.
Therefore, in parallel to increasing the number of parts available and characterising them experimentally, a logical extension to the Registry would be to build a Registry of BioBrick models to complement the physical parts.
A key aspect in this effort is the use of a description language able to describe and support the BioBrick concepts of modularity and abstraction.
| Properties needed for BioBrick description language | |
|
|
Results
In this article, we demonstrate that such a Registry of BioBrick Models is achievable. A mock-up is provided based on the great flexibility and modularity offered by CellML.
Following the steps of already succesful model registries such as the CellML registry or BioModel registry, a BioBrick Model Registry will enable the curation of models. Using CellML and a MIRIAM annotation scheme will guarantee compliance with the previously cited registries. However, a strong emphasis is made on coupling the DNA BioBrick characterisation with their corresponding models. An iterative process between qualitative modelling and experimental characterization will insure consistency. The proposed framework could be the foundation of a future CAD environment for Synthetic Biology.
| Generic CellML architecture for BioBricks | |
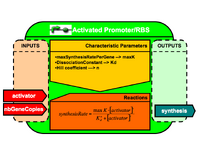 |
First, we explore the definition of modular and re-usable models to represent the available DNA BioBricks. A series of generic model architectures in CellML is defined for most of the types of parts encountered in the DNA registry (plasmid, promoter, RBS, proteins, riboswitch etc.).
Interfaces and import mechanisms in CellML enable a modular and re-usable design. |
| Catalog of quantitative BioBrick models | |
| Second, a catalog of quantitative models based on already characterized parts is presented.
An ongoing effort to characterize BioBricks experimentally is providing us data to move from a qualitative description to a more quantitative one. | |
| Building simulations from modular BioBrick models | |
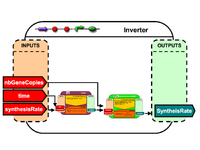 |
To conclude, the versatility of the approach is demonstrated by simulating different systems from a set of pre-defined models: |
Conclusion
The concept of a Registry of BioBrick models based on CellML has been demonstrated. It takes advantage of CellML flexibility and modularity to provide a catalog of quantitative models which are standardized, modular and re-usable. With the increase of available physical DNA parts in the MIT Registry, as well as the characterisation of these parts, such a repository will help to provide a deeper understanding of the BioBrick properties and speed up the process of building new devices and systems. But more importantly, it will help to federate the growing number of contributions from the modeling community and build on the experimental characterization of BioBricks.
