BioSysBio:abstracts/2007/Olga Vasieva
- Add or delete the sections that you require.
Unraveling functional networks: does gene clustering have a meaning?
Author(s): Olga Vasieva
Affiliations: BRC, University of Dundee ; FIG
Contact:email: o.o.vasieva@dundee.ac.uk
Keywords: 'network reconstruction' 'functional prediction' 'gene clustering' 'pathways'
Background/Introduction
Prediction of the role of one protein in a network of regulatory and metabolic interactions involves analysis of many complex data sets in their interference, as no experimental source along is completely and generally reliable. To discriminate the most promising pathways related to a protein of interest from background noise we integrate multiple bioinformatics data by means of a graphical designer/ database of protein-protein interactions supported by data mining and logical modeling of resulted networks. Here we present procedures of unraveling the functional connections between sirtuin family proteins [1] and NF-kappaB regulatory network [2], and the GSTPi [3] function as a carrier of Nitric oxide and Iron complex in the regulation of mitochondrial respiration.
In a process of a network reconstruction we have been evaluating predictive capacities of three data sources: gene expression data, potential protein-protein interaction data derived in two-hybrid system experiments and gene positional clustering [4,5] to show that gene co-localization on a chromosome can be a reliable indication for a functional relevance between the encoded proteins.
We demonstrate a capability of the SEED database and its unique tools for eukaryotic chromosome loci alignment and phylogenetic analysis to serve in retrieval of conservative gene patterns and uncovering of potential functional partners of a protein of interest.
Images/Tables
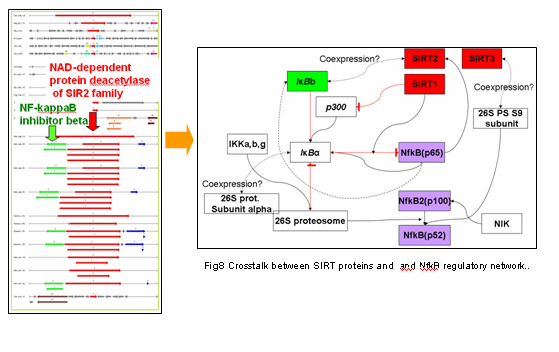
Fig. 1 Shows an example of gene conservative clustering. Chromosome loci (50kb) from different vertebrate genomes are ‘pinned through’ SIRT2 homologs. Arrows correspond to open reading frames: red-SIRT2 homologs, green-NF-kappaB inhibitor beta To the right: A proposed model of interactions between SIRT proteins and and NF-kappaB regulatory network. Dashed lines-putative links derived from the gene positional clustering.
Materials/Methods
The SEED database [1] is an open-source genomic platform provided by the Fellowship for Interpretation of Genomes (FIG) [2], which supports the encoding and projection of metabolic subsystems across the entire collection of integrated genomes. It provides two similar but distinguished approaches which were applied to compare chromosome regions. The first method ‘Compare region’ is similar to one applied to analysis of clustering of functionally related genes on the prokaryotic chromosome [4,6]. The approach involves computation of "pairs of close bidirectional best hits. Using these pairs, one can compose evidence (based on the number of distinct genomes and the phylogenetic distance between the orthologous pairs) that a pair of genes is potentially functionally coupled. Another approach underlies ‘Pinned regions’ resource. It allows one to align chromosome loci that contain open reading frames for homologous proteins, or, in other words: to ‘pin’ these loci through homologous genes. Similarity threshold can be customized. Gene Spring was used for an initial analysis of micro array data; Seed and NCBI databases-for the analysis of sequences derived from Two-Hybrid experiments. All data were integrated by means of PathBlazer[3].
References
1.Smith JS et al.:A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family, Proc Natl Acad Sci U S A,.2000, 97(12):6658-63
2.Giardina C, Hubbard AK : Growing old with nuclear factor-kappaB. Cell Stress Chaperones.,7(2):207-12, 2002
3.Ruscoe JC etal: Pharmacologic or genetic manipulation of glutathione S-transferase P1-1 (GSTpi) influences cell proliferation pathways. J Pharmacol Exp Ther. 2001 Jul;298(1):339-45
4.Overbeek R, et al.: The use of gene clusters to infer functional coupling. Proc Natl Acad Sci U S A., 1999, 96(6):2896-901
5.Lee JM, Sonnhammer EL: Genomic gene clustering analysis of pathways in eukaryotes. Genome Res., (5):875-82, 2003.
6.Overbeek R, et al.: The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005 Oct 7;33(17):5691-702
For further help on editing, please see [ http://en.wikipedia.org/wiki/Help:Editing here]