BioSysBio:abstracts/2007/Guillermo Rodrigo
iGEM 2006 Valencia: EcoliTaster
Author(s): G. Rodrigo1, P. Tortosa3, A.
Aparici2, MC. Aroca2, J. Carrera1, C. Edo1,2, G. Fuertes2, D. Jiménez2, C. Mata2, JV. Medrano2, A.
Montagud2, C. Navarrete2, E. Navarro1, M. Báguena1, P. Fernández de Córdoba1, A.
Ferrando2, J. Salgado2, J. Urchueguía1,A. Jaramillo3
Affiliations: 1Universidad Politecnica Valencia, Spain
- 2Universidad de Valencia, Spain
- 3Ecole Polytechnique, France
- 2Universidad de Valencia, Spain
Contact: http://www.enseignement.polytechnique.fr/profs/biochimie/Alfonso.Jaramillo/
Keywords: 'iGEM' 'Synthetic Biology' 'Computational Protein Design'
Background/Introduction
Our project for this iGEM edition consists in making a cellular biosensor. We use E. coli as cellular chassis, using a deficient EnvZ strain. We construct two different devices (a sensor and an actuator) that are assembled using OmpR-P as a mediator. The sensor device contains a receptor protein and a synthetic two-component signal transduction pathway. The receptor protein has been designed computationally using a ribose binding protein as a scaffold and finding the necessary mutations to change its ligand affinity towards a vanillin molecule. The actuator device takes as input OmpR-P and as output GFP and RFP.
Our sensor device has been inspired on Hellinga’s work sensing TNT and other molecules using a mutated periplasmic binding protein (PBP) [2]. Thus, our team thought in building a PBP that docks a vanillin molecule. It performs an allosteric motion that makes it binding to the trg protein (Fig. 1). When the PBP-vanillin complex (Fig. 2) binds trg, then an allosteric motion is propagated to the EnvZ kinase domain resulting in autophosphorylation and phosphate transfer to OmpR transcription factor (OmpR-P).
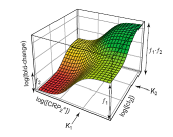
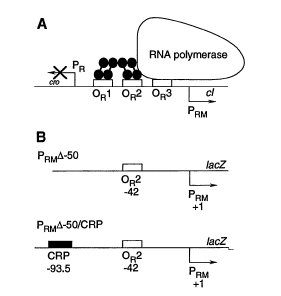
We use a genetic synthetic network as actuator, which at high input levels has a given fluorescent response and at low levels other (Fig. 3). Therefore, for intermediate levels there is a gradient if we superpose the colors. To get that behavior, we construct two promoters with different strengths, and we use a synthetic promoter synergistically activated by two transcription factors cI and CRP [3]. In addition, this promoter can be understood as an AND logic gate (see promoter behavior in Fig. 4, and see its construction in Fig. 5).
Results
We have created several new parts, in particular the CDS of a vanillin receptor, the AND promoter, the trg-EnvZ fusion protein will be very useful to other synthetic biology projects. Our sensor device can be used to construct other cellular biosensors provided the CDS part of our redesigned rbsB is replaced. Our AND promoter will allow to increase the complexity of synthetic circuits constructed from biobricks by using two-regulator promoters. This increase in complexity will permit to reduce the size of genetic circuits performing a given function.
Images/Tables
Materials/Methods
We have used DNA synthesis to obtain the CDS of the vanillin receptor. For the synthesis of the AND promoter we have used DNA synthesis from oligos. The CDS for the CRP and trg where obtained from genomic PCR using DH5a.
For more details, please visit this link.
Conclusion
We have designed a genetic system consisting on few genes that it is expected to give a graded response according to vanillin concentration. This will lead to pattern formation as in the work of Weiss. Our use of a two-regulator promoter allowed to reduce the number of genes required for our actuator device.
References
[1] http://parts.mit.edu/wiki/index.php/UPV-UV_Valencia%2C_Spain_2006
[2] L.L. Looger, M.A. Dwyer, J. Smith, and H.W. Hellinga. Computational design of receptor and sensor proteins with novel functions. Nature, 423, 185-190 (2003).
[3] J.K. Joung, D.M. Koepp, and A. Hochschild. Synergistic activation of transcription by bacteriophage lambda cI protein and E. coli cAMP receptor protein. Science, 265, 1863-1866 (1994).