Berkmen

Bio
Melanie Barker Berkmen
Professor of Chemistry and Biochemistry
Chair of the Division of Physical Sciences (Chemistry and Biochemistry, Physics, Environmental Science and Environmental Studies)
Suffolk University
How to contact me:
Email: mberkmen at suffolk.edu
Office Phone: 617-973-5321
Mailing address:
Suffolk University
Department of Chemistry and Biochemistry
8 Ashburton Place
Boston, MA 02108
Campus Address:
Office: Samia Center (20 Somerset), Room 827
Research Lab: Samia Center (20 Somerset), Room 818
Courses Taught
CHEM 106 and CHEM H106 (Biotechnology and its Applications in Medicine, Agriculture and the Law)
CHEM L111 (General Chemistry Lab I)
CHEM L112 (General Chemistry Lab II)
CHEM 331 (Biochemistry I)
CHEM 332 (Biochemistry II)
CHEM L331 (Biochemical Techniques Lab)
CHEM L332 (Advanced Biochemical Techniques and Research Lab), former iterations known as CHEM L432 and L433
CHEM 428 (Research & Seminar I)
CHEM 429 (Research & Seminar II)
CHEM 510 (Independent Study)
Education
(2002-2007) Jane Coffin Childs Postdoctoral Fellow
Massachusetts Insitute of Technology, Cambridge, MA
Laboratory of Alan D. Grossman
(2001) Ph.D., Cellular and Molecular Biology
University of Wisconsin-Madison, Madision, WI
Laboratory of Richard L. Gourse
(1995) B.S., Biochemistry
University of Dayton, Dayton, OH, summa cum laude
Research
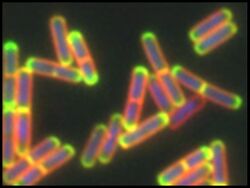
Bacteria have the remarkable ability to acquire new genes in a process known as mating, or conjugation. Mating has played a profound role in bacterial evolution by spreading genes that allow bacteria to adapt to new environments or gain resistance to various antibiotics. During conjugation, DNA is transferred from one cell to another through a specialized translocation channel in the membrane. Many of the molecular mechanisms behind the conjugation process remain a mystery. My research group focuses on characterizing ConB and ConE, two critical interacting protein components of the conjugation machinery of the model bacterium Bacillus subtilis. Our research uses a combination of bioinformatics, molecular, cellular and biochemical approaches to provide insight into how these two proteins function, interact, and localize within the cell. As these proteins are conserved, our findings will apply to the conjugation machinery of numerous Gram-positive bacteria, including many important human pathogens, and provide a deeper understanding of a major mechanism mediating horizontal gene transfer. Our research has been funded through Suffolk University and NSF-RUI grants.
Collaborators
Dr. Alan Grossman, Professor and Chair of Biology, MIT [1]
Dr. Paul Kasili, Professor and Director of Biotechnology Program, Bunker Hill Community College [2]
Dr. Todd Weaver, Professor of Chemistry and Biochemistry, University of Wisconsin-La Crosse [3]
Current Members of the Berkmen Lab
Marcela Melendez - Biochemistry Major, McNair Scholars, May 2023
Marcela joined the lab in the summer of 2021. She learned a variety of techniques including the preparation and sterilization of media, mating assays, and DNA purification. She determined how mitomycin C concentration affects mating frequency.
Jillian Allen - Biochemistry Major, Honors, May 2023
Jillian joined the lab in the summer of 2022. She learned a variety of techniques including the preparation and sterilization of media and fluorescence microscopy. She analyzed what proteins ConB interacts with using bacterial-two-hybrid.
Abby Thornhill - Biochemistry Major, Honors, May 2023
Abby joined the lab in the summer of 2022. She learned a variety of techniques including the preparation and sterilization of media and bacterial two-hybrid assays. She analyzed ConB subcellular localization using fluorescence microscopy.
Alexandria Russo - Biology Major, Honors, May 2024
Alex joined the lab for the summer of 2022. She learned a variety of techniques including the preparation and sterilization of media, bacterial-two-hybrid, and DNA purification and quantitation.
Former Members of the Berkmen Lab



Hunter Toyoda Biochemistry major, Honors, May 2022
Hunter joined the lab in the spring of 2021. He mastered a variety of techniques including bacterial two hybrid assays and Q5 mutagenesis. He investigated whether mutations in ConE affect its interaction with itself and the ConB protein using bacterial two hybrid studies.
Misael Flores Biochemistry major, May 2022
Misael joined the lab in the summer of 2021. He mastered a variety of techniques including the preparation and sterilization of media and mating assays. He explored how DNA damaging reagents affect mating frequency.
Kathy Nguyen Biochemistry major, Honors, May 2022
Misael joined the lab in the summer of 2021. She mastered a variety of techniques including bacterial two hybrid assays and Q5 mutagenesis. She analyzed how mutations in ConB affect protein stability using quantitative western blots.
Olukemi Akinleye - Biochemistry Major, Honors program, May 2021
Olukemi joined the lab in the summer of 2018. He learned a variety of techniques including the preparation and sterilization of buffers and media, preparation of competent cells, bacterial transformation, mating assays, agarose gel electrophoresis, and gel shift assays. He specialized in generating models of proteins for 3D printing and projection in augmented reality visors. He analyzed the kinetics of mating and mobilization.
Sirui Chen - Biochemistry Major, Honors program, May 2021
Sirui joined the lab in the summer of 2019. He learned a variety of techniques including the preparation and sterilization of buffers and media, preparation of competent cells, bacterial transformation, and mating assays. He specialized in fluorescence microscopy. He explored the localization of ConB and ConE.
Adelyn Ragucci - Biochemistry Major with minor in Forensic Science, Honors program, May 2020
Adelyn joined the lab in the summer of 2018. She mastered a variety of techniques including the preparation and sterilization of buffers and media, preparation of competent cells, bacterial transformation, mating assays, agarose gel electrophoresis, gel shift assays, site-directed mutagenesis, protein purification and protein characterization. She used bacterial two-hybrid to study the oligomerization and interactions between the mating proteins ConB and ConE.
Cristelle Badaoui - Biology Major, Honors program, May 2020
Cristelle joined the lab in the summer of 2019. She learned a variety of techniques including the preparation and sterilization of buffers and media, preparation of competent cells, and bacterial transformation. She used bacterial two-hybrid to study the oligomerization and interactions between the mating proteins ConD and itself, ConB, and ConE.
Haley Dame - B.S. Biochemistry, Honors Program, May 2019
Haley joined the lab in the summer of 2017. She picked up a number of techniques including protein purification and characterization. For her senior thesis, she characterized the location of ConB within B. subtilis cells. For this project, she cloned a number of fluorescent proteins and tags to the conB gene and visualized them using fluorescence microscopy.
Betelhem Gemechu - B.S. Biochemistry, May 2019
Betelhem joined the lab in the summer of 2017. She has learned a variety of microbial and biochemical techniques. For her thesis, she explored the kinetics and genes required for mating, plasmid mobilization, and retro-transfer.
Sabrina Ali - B.S. Biology, Honors Program, May 2019
Sabrina joined the lab in the summers of 2016 and 2017. She learned a variety of microbiological and molecular biology techniques. Her project was focused on characterizing the determinants of mating and retro-mobilization of ICEBs1.
Meri Kalashyan - B.S. Biochemistry, Honors program, May 2018
Meri joined the lab in the summer of 2017. She used a variety of biochemical and microbiological techniques to study the levels of different ICEBs1 mating proteins inside B. subtilis cells. She is a master of the quantitative western blot using infrared fluorescence.
Kacie McCarty - B.S. Biochemistry with concentration in Forensic Science, May 2018
Kacie joined the lab in the summer of 2017. She used bioinformatics, biochemistry and some molecular biology to study ConQ, the putative coupling protein of ICEBs1. She has become a master of nickel affinity chromatography, protein purification and protein characterization.
Sabrina Phanor - B.S. Biochemistry, Honors and McNairs Scholars Programs, May 2018
Sabrina joined the lab in the summer of 2015. She learned fluorescence microscopy and site-directed mutagenesis. She also purified and characterized His6-tagged ConB truncations. Finally, for her senior thesis, Sabrina biochemically characterized two clinically relevant mutations in the human fumarase gene. She introduced the mutations, purified the proteins, and kinetically characterized them.
Sydney Thomas - B.S. Biochemistry, McNairs Scholars Program, May 2018
Sydney joined the lab in the Fall of 2015. She used site-directed mutagenesis techniques to create several new conB constructs lacking various conserved domains. She has also used Gibson (Isothermal) assembly and traditional cloning to construct various sumo-tagged proteins.
Michelle Lopez - B.S. General Studies, McNair Scholars Program, May 2017
Michelle joined the lab in the summer of 2016. She learned a variety of microbiological, molecular and biochemical techniques and discovered which domains of ConB are required for mating.
Artemisa Bulku - B.S. Biochemistry, Honors Program, May 2016
Artemisa joined the lab in the summers of 2012 and 2013. She cloned His10-ConE, along with several mutants of ConE. In addition, she has optimized the purification of His10-ConE, providing a better yield and purity for antibody production. She also started a whole new project on fumarase for the Advanced Biochemistry Research lab.
Naira Aleksanyan - B.S. Biology, McNairs Scholars Program, December 2015
Naira joined the lab in the summer of 2014. She created several site-directed mutants of ConE and examined their effects on mating and showed that their levels were normal inside cells.
Anastasia Murthy - B.S. Biochemistry, Honors Program, May 2015
Anastasia joined the lab in the summer of 2013. After that, she explored the localization of several ConE mutants along with optimizing the purification of MBP-tagged ConE. Anastasia won a prestigious NSF Graduate Fellowship and is pursuing a PhD in Molecular Biology, Cell Biology, and Biochemistry at Brown Unviersity.
Omar Pinkhasov - B.S. Biochemistry, Honors Program, May 2015
Omar joined the lab in the summer of 2014. He purified full length ConB and a truncated version lacking its transmembrane segments, and explored the oligomerization of these proteins using Blue Native PAGE.
Matthew Broulidakis - B.S. Biochemistry, Honors Program, May 2014
Matthew joined the lab in the summer of 2012. He explored the role of yddF in mating. Then, he examined the determinants of localization of ConE-GFP as well as analyzed the localization of other mating components through GFP and SNAP tags. After graduation, Matthew became an associate scientist at Pfizer.
Minh Bui - B.S. Biochemistry, May 2014
Minh constructed 4 different ConE mutations on the chromosome and analyzed their effects on mating for her senior thesis. After graduation, Minh returned to Vietnam and is now working as a research technician.
Kyle Swerdlow - B.S. Biochemistry, May 2014
Kyle worked very closely with Gianna Mancuso on the optimization of His6-ConE purification. They presented this research at the 2012 ACS national conference as well as the 2012 STEM conference at Suffolk University. Funded through the NSF-RUI grant during the summers of 2012 and 2013, Kyle used bacterial two-hybrid analysis to piece together key players in the ICEBs1 conjugation machinery for his senior thesis.
Gianna Mancuso - B.S. Biochemistry Forensic Science, May 2013
Gianna, along with Kyle Swerdlow, helped optimize the purification of His6-ConE. The research was presented at the 2012 ACS national conference and 2012 STEM conference at Suffolk University. She has also helped clone ConE fusions for use in bacterial two-hybrid. After graduation, Gianna became a tissue-typing technologist at Brigham and Women's Hospital in Boston.
Georgeanna Morton - B.S. Biochemistry, May 2013
Geogeanna joined the lab in the summer of 2012. For her senior thesis, she purified wild-type and mutant His6-ConE and tested the oligomerization of each using Blue Native-PAGE. After graduation, Georgeanna became a research assistant at Beth Israel Deaconess Medical Center in Boston.
Azul Pinochet Barros - Biology/Philosophy, December 2012
Azul helped optimize our competent cells procedure. Afterwards, she cloned a Walker B mutant for our His6-ConE, but unfortunately this mutant did not express well in E. coli. After graduation, Azul entered a PhD program in microbiology at Cornell University.
Stephanie Laurer - B.A. Biochemistry, Honors Program, May 2012
Stephanie used mating assays to determine that addition of a His6-tag on the N-terminus of ConE does not interfere with ConE's ability to support mating. She presented this research with Bridget at the 2010 ACS National Meeting in San Francisco and at the Suffolk Science Banquet where they won a poster award. She also helped clone several ICEBs1 genes. On the side, Stephanie had an interest in genetically modified food. During her first year at Suffolk, she used a PCR-based assay to detect the Bt gene in corn. She found that at least half of the samples she tested (6 of 11) were genetically modified. She presented this work at the 2009 Suffolk science banquet where her poster won first place. After working as a Clinical Research Assistant at Beth Israel Deaconess Medical Center, she entered a Physician's Assistant graduate program at Northeastern University and is now a Physician Assistant.
Bridget Giarusso - B.S. Biochemistry, May 2011
As a research assistant, Bridget used mating assays to determine that addition of a His6-tag on the N-terminus of ConE does not interfere with ConE's ability to support mating. She presented this research with Stephanie at the 2010 ACS National Meeting in San Francisco and at the Suffolk Science Banquet where they won a poster award. She helped clone several ICEBs1 genes. She is now a Clinical Research Assistant at Beth Israel Deaconess Medical Center.

Matt Hamada - B.S. Biochemistry, December 2010
For CHEM L333, Matt cloned the conC gene encoded on the ICEBs1 conjugative element. For CHEM L428/L429, Matt used fluorescence microscopy to determine whether conC and other ICEBs1 genes are required for ConE to localize at the cell poles. He presented his research with Cori at the 2009 Boston Bacterial Meeting. Currently, Matt is working as a laboratory research technician at Midwestern University.
Cori Leonetti - B.S. Biochemistry, May 2010
For CHEM L333, Cori helped clone the conB gene encoded on the ICEBs1 conjugative element. Cori extended her CHEM L333 project for CHEM L428/L429. She tested whether conB and other ICEBs1 genes are required for mating. Cori presented her research with Matt at the 2009 Boston Bacterial Meeting. Cori obtained a Masters in microbiology at Arizona State University and is now teaching at Phoenix College.
Erin Cross - B.S. Biochemistry, May 2009
In CHEM L333, Erin and her class mates attempted to clone various C-terminal truncations of ConE fused to GFP. For CHEM L428/L429, she used fluorescence microscopy to analyze what parts of ConE are required for localization to the cell poles. She found that the C-terminal half of ConE is critical for localization. She presented this work at the national ACS meeting in March 2009 and at the 2009 Suffolk Science Banquet where her poster won 3rd place. Erin is now working as a laboratory research technologist at Maine Medical Center Research Institute.
Maria Muccioli (formerly Levicheva) - B.S. Biochemistry, Honors Program, May 2009
For CHEM L333, Maria constructed a his-tagged ConE. For CHEM L428/L429, she purified and characterized His6-ConE to enable future students to perform ATPase assays to determine whether ConE can hydrolyze ATP in vitro. She presented this work at the national ACS meeting in March 2009. Maria obtained a PhD in molecular and cellular biology from Ohio University and is now a post-doc at Ohio State University.
Tamara Wong - B.S. Biochemistry Forensic Science, May 2009
Tamara helped cloned the His6-ConE construct so that we can test whether this protein can support mating. In addition, Tamara purified His6-ConE to enable future ATPase assays. Tamara is now a senior Forensic Scientist for the State of Rhode Island.
Emma-Kate Loveday - B.S. Biochemistry, May 2008
For her CHEM L428/L429 project, Emma-Kate constructed two variants of ConE and tested their effects on mating. She found that the Walker B (ATP hydrolysis domain) of ConE is essential for mating. She also found that the N-terminus of ConE does not contribute significantly to mating. She presented her work at the Boston Bacterial Meeting in June 2008 and the Cold Spring Harbor Molecular Genetics of Bacteria and Phages Meeting in August 2008. Emma obtained a PhD in microbiology and immunology at the University of British Columbia supported by an NSF Fellowship.
Publications
Suffolk undergraduate student co-authors in red
Peterson CN, Tavana SZ, Akinleye OP, Johnson WH, Berkmen MB. (2020) An idea to explore: Use of augmented reality for teaching three-dimensional biomolecular structures. Biochemistry and Molecular Biology Education, 1-7.
Grocott O, Phanor SK, Fung F, Thibert RL, Berkmen MB. (2019) Clinical report and biochemical analysis of a patient with fumarate hydratase deficiency. American Journal of Medical Genetics Part A, 182A:504-507.
Bulku A, Weaver TM, Berkmen MB. (2018) Biochemical characterization of two clinically-relevant human fumarase variants defective for oligomerization. The Open Biochemistry Journal, 12:1-15.
Auchtung JM, Aleksanyan N, Bulku A, Berkmen MB. (2016) Biology of ICEBs1, an integrative and conjugative element in Bacillus subtilis. Plasmid, 86:14-25.
Leonetti CT, Hamada MA, Laurer SJ, Broulidakis MP, Swerdlow KJ, Lee CA, Grossman AD, Berkmen MB. (2015) Critical components of the conjugation machinery of the integrative and conjugative element ICEBs1 of Bacillus subtilis. Journal of Bacteriology, , 197(15): 2558-67.
Berkmen MB, Murthy AC, Broulidakis MP. (2014) An Inquiry-Based Laboratory Module to Promote Understanding of the Scientific Method and Bacterial Conjugation. J Micriobiol & Biol Educ., 15(2): 321-2.
Berkmen MB, Laurer SJ, Giarusso BK, and Romero R. (2013) The Integrative and Conjugative Element ICEBs1 of Bacillus subtilis. Review chapter in the book Bacterial Integrative Mobile Genetic Elements, edited by Adam P. Roberts and Peter Mullany, Landes Bioscience.
Martinez KA 2nd, Kitko RD, Mershon JP, Adcox HE, Malek KA, Berkmen MB, Slonczewski JL. (2012) Cytoplasmic pH response to acid stress in individual cells of Escherichia coli and Bacillus subtilis observed by fluorescence ratio imaging microscopy. Appl Environ Microbiol., 78(10):3706-14.
Babic A, Berkmen MB, Lee CA, Grossman AD. (2011) Efficient gene transfer in bacterial cell chains. MBio, 2(2). pii: e00027-11. doi: 10.1128/mBio.00027-11.
Berkmen MB, Lee CA, Loveday EK, Grossman AD. (2010) Polar positioning of a conjugation protein from the integrative and conjugative element ICEBs1 of Bacillus subtilis. J Bacteriol, 192(1):38-45.
Kitko RD, Cleeton RL, Armentrout EI, Lee GE, Noguchi K, Berkmen MB, Jones BD, Slonczewski JL. (2009) Cytoplasmic acidification and the benzoate transcriptome in Bacillus subtilis. PLoS One, 4(12):e8255.
Vrentas CE, Gaal T, Berkmen MB, Rutherford ST, Haugen SP, Ross W, Gourse RL. (2008) Still looking for the magic spot: the crystallographically defined binding site for ppGpp on RNA polymerase is unlikely to be responsible for rRNA transcription regulation. J Mol Biol, 277(2): 551-64.
Wang JD, Berkmen MB, Grossman AD. (2007) Genome-wide co-orientation of replication and transcription reduces adverse effects on replication in Bacillus subtilis, PNAS, 104(13): 5608-5613.
Berkmen MB and Grossman AD. (2007) Subcellular positioning of the origin region of the Bacillus subtilis chromosome is independent of sequences within oriC, the site of replication initiation, and the replication initiator DnaA. Mol Microbiol, 63(1): 150-165.
Berkmen MB, Grossman AD. (2006) Spatial and temporal organization of the Bacillus subtilis replication cycle. Mol. Microbiol, 62(1): 57-71.
Haugen SP, Berkmen MB, Ross W, Gaal T, Ward C, Gourse RL. (2006) rRNA promoter regulation by nonoptimal binding of σ region 1.2: An additional recognition element for RNA polymerase. Cell, 125(6): 1069-1082.
Paul BJ, Berkmen MB, Gourse RL (2005) DksA potentiates direct activation of amino acid promoters by ppGpp. PNAS, 102(22):7823-8.
Paul BJ, Barker MM, Ross W, Schneider DA, Webb C, Foster JW, Gourse RL (2004) DksA: A critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell, 118(3): 311-322.
Wang JD, Rokop ME, Barker MM, Hanson NR, Grossman AD (2004) Multi-copy plasmids affect replisome positioning in Bacillus subtilis. J Bacteriol, 186(21):7084-90.
Barker MM, Gourse RL (2002) Control of stable RNA synthesis. In Translation Mechanisms. (Lapointe J, Brakier-Gingras L. ed.). Landes Biosciences, Austin, TX.
Barker MM, Gourse RL (2001) Regulation of rRNA transcription correlates with nucleoside triphosphate sensing. J Bacteriol, 183, 6315-6323.
Barker MM, Gaal T, Josaitis CA, Gourse RL. (2001) Mechanism of regulation of transcription initiation by ppGpp. I. Effects of ppGpp on transcription initiation in vivo and in vitro. J Mol Biol 305(4): 673-688.
Barker MM, Gaal T, Gourse RL (2001) Mechanism of regulation of transcription initiation by ppGpp II. Models for positive control based on properties of RNAP mutants and competition for RNAP. J Mol Biol 305(4): 689-702.
Gourse RL, Gaal T, Aiyar SE, Barker MM, Estrem ST, Hirvonen CA, Ross W. (1998) Strength and regulation without transcription factors: Lessons from bacterial rRNA promoters. Cold Spring Harb Sym 63: 131-139.
Personal
In my free time I like to travel and cook.