Berk2006-ConjugationTeam
Return to main page
SIL_07/21/06
So, the knockout plates for the F plasmid came back as lawns that didn't even smell like e.coli (according to john) so they are definitely not what we were going for. Our guess with what went wrong is that pOX38 may be CAM resistant, which would be very bad, so now we're streaking some F plasmids, in particular Frost F, and hopefully one of them will be CAM sensitive so that we can do the knockout.
On the other hand, the R plasmid knockout of traG looks promising, so Matt is growing overnight cultures of 4 clones and tomorrow we will do a genomic mini-prep and map it tomorrow to see if the gene is knocked out.
Also, the complements grew last night, so I mini-prepped them all today. One of the TraM complement clones came out pink which indicates it is probably the parent vector so I did not mini prep that one. I also digested them in EcoR1/Spe1 and ran them on a gel to check to see if the insert is around the right size. Also we are going to send them in for sequencing.
Picture of complements digested in EcoR1/Spe1 (pSB1A2-J23013 (1), pSB1A2-J23013(2), pSB1A2-J23011, ladder)
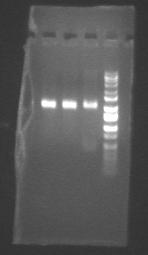
We are also almost out of TriK plates and Carb plates, so Matt poured a bunch more.
MF_07/20/06
Today we are doing the knockouts of pKD46/pOX38 (OriT and trbC knockouts, the TraM PCR and oligos failed) and pKD46/Rlambda (traG knockout). We are going to electroporate the PCR products into competent cells, which we also made today.
Yesterday, we redid the PCR for the OriT knockout using higher concentrations of the oligos, hoping for better results and we got them this morning and the mapping came out pretty good. We are going to use the PCR from the middle lane because it came out cleaner. We cleaned the PCR product, digested it in dPN1 and cleaned it up again. Here is the gel picture:
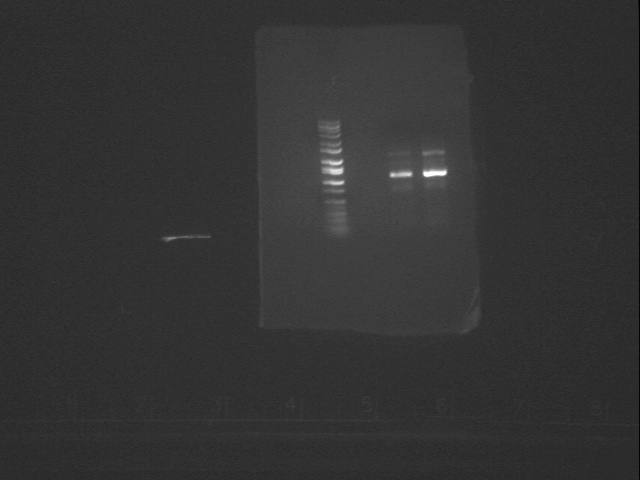
We also grew up overnights of the TraM and TraG biobricks (pSB1A2-J23011 and pSB1A2-J23013), so that tomorrow we can digest them in EcoR1 and Spe1 and map them to see if the fragments are around the right size, and then we will sequence them to make sure.
Lastly, we are hopefully fininshing up the dilution series of the conjugation experiments today, we are going to do several dilution for the 3 cultures that still grew lawns or else grew nothing because of too much dilution.
SIL_07/19/06
Conjugation experiments with EC100D::pir+ as the recipient from 7/18/06
Number of Titer Colonies for EC100D::pir+ = 4
| Culture | OriT | TraJ | # of Colonies on TriK | # of Colonies on TriA | Dilution Factor | # of Titer Colonies |
|---|---|---|---|---|---|---|
| WT pOX38 | + | + | 850 | n/a | 1000 | 52 |
| pOX 38+DH10B/J01064 | + | + | 2880 (100 fold) | 1760 (100 fold) | 100 | 3 |
| Olambda+1064 | + | + | 6 (2 red, 4 white) | 10 (red) | 10 | n/a |
| TlambdaOlambda+1064 | + | - | 50 | 3 | 10 | n/a |
| TlamdaOlamda+1064+1093 | + | + | 1 | 0 | 10 | n/a |
| RP4 | + | + | 30 (10 fold) | n/a | 10 | 38 |
| BW20767+JO1003 | + | + | n/a | 625 | 1000 | 3 |
| Rlambda+JO1003 | + | - | 45 (1:1) | 1280 (1:1) | 0 | 16 |
| Rlambda+JO1003+J01024 | + | - | 550 | 0 | 10 | 2 |
Picture of KOs and Complements PCR gel: (lane 1= ladder, lane 2= sub trbCf, lane 3 = sub traMf, lane 4 = sub traGr, lane 5 = KO OriTf, lane 6 = KO trbCf, lane 7= KO traMf, lane 8 = KO traGr)
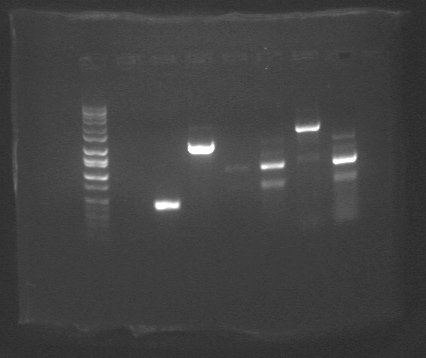
Because of poor mapping results, am redoing subtrbCf, KO OriTf, and KO traMf with Phusion PCR kits, am also rdoing sub trbCf using the Extend kit, but this time changing the annealing temperature to 45 degrees, and also adding 10% DMSO to one of 2 Extend PCR preps of it. Here is the gel.
Picture of the gel: (lane 1= subtrbCf extend, lane 2 = subtrbCf extend+dmso, lane 3 = subtrbCf phusion, lane 4 = KO OriTf phusion, lane 5 = KO traMf phusion, lane 6= ladder)
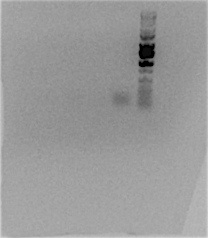
All of them failed.
The ones that had good mapping results, we cleaned up the PCR preps and are digesting them with dNP1 to cut all the methylated products, and then cleaning them again for future use.
Also, pSB1A2-J23012, the SpecR biobrick grew in a AMP/SPEC cultures, so chances are that it is what we think it is, so I'm miniprepping 2 clones of it.
The results from the R plasmid streaking from the -80 revealed that none of our variants were Amp sensitive and Kan resistant. In fact, only PCV was Amp sensitive, and it was Cam resistant. So we are just going to do the knockout on R lambda instead of something new.
Today, we also made pSB1A2-J23011 which is the TraMf biobrick and pSB1A2-J23013 which is the TraGr biobrick, from the PCR digests.
SIL_07/18/06
We need to find an Amp sensitive variant of RP4, so we streaked a bunch of entries in the -80 that could have been it: PK3, PK2, RP4 (Amp,Kan,tet), RP1xK1200Kan, RP1xK1200ND, RP4 (Amp,Kan,Tet), RP1xK1200Amp, PCV. We streaked them onto Amp, Kan, and Cam plates and also put them into 5 ml of LB to grow up overnight. Also, just in case none of these variants were good, we went ahead and transformed Rlambda with pKD46 (two times, once with a rescue, and one without) and put them in the 30 degree incubator.
We are also redoing the conjugation experiment for the plates that had too many colonies to count, with an added dilution step at the end and also titering right before the mating reaction. We are going to titer by diluting by 6 orders of magnitude (1uL into 1mL, then 1uL into 1mL again)and then plating it on 50 uL.
Lastly, we are doing Knockout PCRs for the F plasmid, which are ForlamF/R on pkD3, trC-F on pOX38 using JL12 and JL13, and the traM-F on pOX38 using JL3 and JL7. We are also doing PCRs for the subs of the knockouts, so that we can make new biobrick complements: trbC using JL14 and JL15, traMf using JL5 and JL6, and traGr using JL10 and JL11.
The traG knockout will be done using JL8 and JL9 once we find a variant of RP4 that isn't Amp resistant to use.
JL_07/17/06
conjugation experiments with EC100D::pir+ as the recipient
| Culture | OriT | TraJ | # of Colonies on TriK | # of Colonies on TriA | Comments |
|---|---|---|---|---|---|
| RP4 | + | + | ~1000 | n/a | |
| BW20767+JO1003 | + | + | n/a | lawn | |
| Rlambda+JO1003 | + | - | 45 | >1000 | |
| Olambda+1064 | + | + | 6 (2 red, 4 white) | 10 (red) | |
| TlambdaOlambda+1064 | + | - | 50 | 3 | |
| TlamdaOlamda+1064+1093 | + | + | 1 | 0 | |
| WT pOX38 | + | + | lawn | n/a | |
| pOX 38+J01064 | + | + | lawn (1/3 red, 2/3 white) | lawn (4/5 red, 1/5 white) |
1.Results of RP1xEC100D::pir+ on Amp= lawn (BAD! we do not want RP1 to have Amp resistence)
2.Results of pkD46xDH10B + RP1xEC100D::pir+ on KanAmp= lawn
3.We also made pSB1A2-j23012 which is the Spec resistent biobrick
4.We digested F plasmid and Olambda with NcoI and ran a gel.
Picture of the gel: (lane 1= F plasmid, lane 2= Olambda)
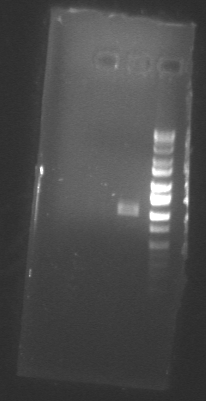
The results are not what we predicted. Olambda should have two bands because it is a linear fragment and was cut once with NcoI
5. We are growing everything that was not countable in the conjugation experiments that we did on 07/15/06
6. We are growing pOX38 mated with EC100D::pir+ and pKD46xDH10B. We are going to mate them tomorrow
MF 07/14/06
We had to re-grow EC100D::pir+ and pOX38 in straight LB so that we could make aliquots with no antibiotic. The reason for this is that it provides a better baseline if all the cells are frozen without antibiotic and it also saves time because we don't have to wash the antibiotics out of the aliquots. pSB1A2-JO1003 with Rlambda-pSB3C6-JO1024 is also growing up in the incubator because it didn't grow overnight. I made aliquots of EC100D::pir+ and pOX38 in glycerol. They are in the cardboard box in the back compartment of the -80.
protocol for conjugation 07/14/06
donor protocol
- start out with saturated 1 mL of culture
- spin down (top speed for 10 sec.) , discard fluid
- resuspend in 1 mL LB
- vortex, spin, discard fluid
- resuspend in 1 mL LB, vortex
- put 200uL of culture into 2 mL of LB (if arabinose is needed put in 100 mg/mL)
- grow 2 hours
- do Assay
Recipient protocol
- take aliquot and thaw
- dilute 10 fold into LB (ie. put 200 uL into 2 mL of LB)
- grow 2 hours
- do Assay
JL&SIL_07/13/06
Today we conjugated RP1xK1200(Kan) with EC100D::pir+ and plated it on TriK. We used RP1 instead of RP4 because RP4 is also amp resistent (which we do not want). Tomorrow we are going to conjugate EC100D+RP1 with DH10B+pKD48 to make aliquots of DH10B+RP1.
Today we also made aliquots of EC100D::pir+ and pOX38 as glycerol stocks to put in the -80 freezer.
Also, the transformation of DH10B and pKD46 in the 30 degree incubator did not grow at all, so we redid the transformation both as a heat shock and also an electroporation today and hopefully it will work this time.
Finally, we are preparing to do a quantitative conjugation experiment with the F plasmids and R plasmids. So I went through a whole bunch of trouble to make overnight saturated cultures of some colonies because we were out of antibiotics so I had to make more and now we have a bunch of Kan stored in a 50 mL tube in the freezer, as well as a bunch of aliquots in a box in the freezer.
Here's the list of cultures:
- pOX38 (K)
- pOX38 + pSB1A2-J01064 (KA)
- Olambda + pSB1A2-J01064 (KA)
- TlambdaOlambda + pSB1A2-J01064 (KA)
- TlambdaOlambda + pSB1A2-J01064 + pSB3C6-J01093 (KAC)
- RP4 (K)
- Rlambda + pSB1A2-J01003 (KA)
- Rlambda + pSB1A2-J01003 + pSB3C6-J01024 (KAC)
- BW20767 + pSB1A2-J01003 (A)
- Note: EC100D:pir+ for these experiments can be found as a glycerol stock in a box in the -80 freezer, as can pOX38 in the future. We are also going to make glycerol stocks of RP1 to put in the freezer.
The oligos that we ordered on 07/11/06 came! Jenn has ordered even more oligos for the TraG knockout and complement for RP1 and oligos for the trbC knockout and complement for pOX38, the F plasmid.
We did a PCR prep for oligos JL1, JL2, JL3, and JL4. For JL1 and JL2 we used the DNA template "psplam". For JL3 and JL4 we used the DNA templates Olambda and F. J1 and J2 are for the SpecR biobrick and J3 and JL4 are for the characterization of the OriTf and TraM region.
SIL_07/12/06
Today the results from the pOX38 x1064 conjugation experiment are recorded in the table from 07/10/06 with the rest of the results. I minipred pSB1A2-J23010 today and mapped it, and it came back positive for both OriTf and OnGFP. Below is a picture of the analytical gel from the mapping.
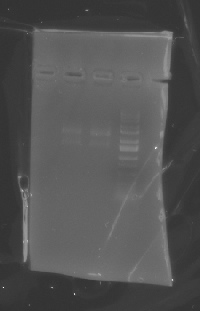 (Map of pSB1A2-J23010)
(Map of pSB1A2-J23010)
Also, today Matt made more competent cells of TGI cells. However, we ran out of liquid nitrogen so we had to put the TGI's into the freezer so that they are going to experience a slow freeze and we don't know how well that is going to work out.
We also transformed pKD46 with DH10B cells onto an Amp plate and we're keeping it in a 30 degree incubator, in order to prepare ourselves to do a knockout with the lambda red plasmids necessary in the reaction.
An important decision was made today about making our experiments quantitative instead of merely qualitative. Thus we need to be consistent in a lot of variables and create standard procedures for conjugation experiments. Thus we are growing up overnight colonies of EC100D and pOX38, so that we can aliquot them tomorrow and create 200 uL glycerol stocks of them to put in the -80 freezer, that we can use for future experiments.
We also restreaked pOX38 onto a Kan plate so that we have some fresh colonies.
Also, we are ordering oligos to isolate the TraG gene in the R plasmid, which is the gene we have decided to control because its removal decreases conjugation activity by about 7 orders of magnitude and can be complemented on another plasmid. However we are still looking for an equivalent gene for the F plasmid. Jenn has been searching for "F plasmid" and "Tra*" papers online for more information.
Right now we are going to mate RP1 (which is in a mystery cell strain and has a mystery plasmid with it) and EC100D on TriK, so that the RP1 plasmid passes into the EC100D cell. Tomorrow, we will mate the RP1 and EC100D cell with the pKD46 xDh10B cell that I transformed today. That will result in a DH10B cell with an RP1 plasmid and a pKD46 plasmid that we will plate on KanAmp at 30 degrees. Using 30% of those cells, we will do RP1 knockouts. The rest we will cure at 43 degrees so that the pKD46 disappears and we are left with only RP1 in DH10B, and we will grow those cells in a culture and make 50% glycerol stocks from them.
MF_07/11/06
Results from the conjugations we performed yesterday are in the table under yesterdays date. We also conjugated pOX38+ JO1064 with EC100d::pit+.
The pSB1A2-J23010 that Sam made yesterday was successful and glows intensely green under UV light. We are currently growing overnight cultures of it, so that we can miniprep it tomorrow and also map it to confirm its identity (have to check that OriT is there, obvious that OnGFP is there).
We are designing oligos to make a TraM bioBrick, a Spec Resistant bioBrick, and also oligos to do knockouts on the wild type F plasmid.
Also...MORE PLATES!
JL_07/10/06
On Sunday, Matt grew up saturated cultures of EC100D::pir+, Rlambda/J01003+1024, BW20767+JO1003, Rlambda+JO1003, TG1+JO1003, WT RP4, Rlambda, WT pOX38.
Today, we performed conjugation experiments with EC100D::pir+ as the recipient.
| Culture | OriT | TraJ | # of Colonies on TriK | # of Colonies on TriA | Comments |
|---|---|---|---|---|---|
| Rlambda/J01003+1024 | + | + | no colonies | >1000 | both expected, 1003 has Amp, 1024 not transfering |
| BW20767+JO1003 | + | + | n/a | lawn | 1003 adds OriT and Amp resistance |
| Rlambda+JO1003 | + | + | no colonies | >1000 | rlambda not transfering (expected) but 1003 transfering a lot on TriA. We expect less transfer without TraJ. |
| TG1+JO1003 | + | - | no colonies | n/a | expected because no conjugation plasmids in TG1 cells. |
| WT RP4 | + | + | >1000 | n/a | expected |
| Rlambda | - | + | no colonies | n/a | expected- no OriT or TraJ |
| WT pOX38 | + | + | >1000 | n/a | expected- no knockouts |
| pOX 38+J01064 | + | + | lawn (almost entirely white, with a few red colonies) | >1000 (half red,half white) | triK should have more red colonies, triA should only have a few red colonies |
Also,pSB1A2-J23010 did not show any flurorescence, so Sam remade it again today. And this time used an ethynium bromide gel instead of syber green so that the bands were a lot clearer and this time, the right insert was cut out for sure.
JL_07/07/06
Results from 07/06/06 pSB1A2-J01003 transformations
- pSB1A2-J01003 into TGIs on Amp: grew efficiently (>1000)
- pSB1A2-J01003 into Rlambda on Amp: 100 (two phenotypes are seen)
- pSB1A2-J01003 and pSB3C6-J01024 into Rlambda on Kan/Carb/Cam: no colonies (BUT its probably because the SOB media was contaminated)
Since no colonies grew for the 'pSB1A2-J01003 and pSB3C6-J01024 into Rlambda on Kan/Carb/Cam' transformation, we are making competent cells of Rlambda/J01003 to tranform them with j01024.
We also made pSB1A2-J23010 today which is [OriTr]+[OnGFP] which will be useful in the future.
Because our mating experiments of Olambda did not turn out the way we predicted, we transformed J01064 into F plasmid (pk38).
Also, we made oligos for TraY and Spec for our next experiment.
SIL_07/06/06
- The results from 7/05/06 F plasmid mating experiment were exactly the same as before, which most likely indicates that OriT wasn't properly knocked out in Olambda. As for the R plasmid mating experiment, the results also came out strange.
- Results from 7/05/06 R plasmid mating experiment: lots of colonies grew on both plates, with both pSB1A2-J01003 from -80 only and pSB1A2-J01003 into BW20767. More grew on the plate with BW20767, which is good, but the fact that there was almost a lawn in the middle of the plate for only J01003 is a bad sign becuase J01003 should not be able to transfer on its own without the R plasmid. It may be a sign that either J01003 is contaminated with Tri resistance or else it is on a plasmid that may exhibit some R plasmid activity. From now on, we will plate the doner on Tri just to make sure it has none of that resistance, and EC100D:pir+ on Kan and Amp to ensure that they all have the resistances that we assume they have during future experiments.
- We also put tough tags onto pSB1A2-J01064, pSB3C6-J01024, pSB1A2-J01001 and pSB3C6-J01093.
- Using finished PCR products from the night before of the Olambda characterization experiment, we mapped the results this morning and they came out weird. We suspect that we will have to create a new knockout strain that will hopefully perform more as we expect them to.
SIL_07/05/06
Results from 7/03/06 F plasmid mating experiment
| Culture | OriT | TraJ | # of Colonies on TriK | # of Colonies on TriA | Comments |
|---|---|---|---|---|---|
| Olambda | - | + | 100 | n/a | not desired, could mean OriT wasn't knocked out completely |
| pSB1A2-J01064 from -80 | + | - | n/a | 0 | good |
| J01064 into Olambda | + | + | 200 (1/4 pink)) | 100 (half pink) | don't want any colonies on the TriK plate, and it's strange that there are pink colonies, and that the colonies on TriA plate aren't all pink. Could mean that Olambda and pSB1A2-J01064 plasmids transfer at the same time. |
Today, we redid the F plasmid mating experiment to confirm results, and also are doing the R plasmid mating experiment for the first time. The R plasmid mating experiment entails mating just pSB1A2-J01003 from -80 with EC100D:pir+ and pSB1A2-J01003 transformed into BW20767 with EC100D:pir+ and plating them both on TriA plates to see if the 1003 plasmid transfers by itself or with the R plasmid with an OriT knockout. We already know for sure that BW20767 will not transfer by itself because this has been confirmed by other people.
We're also analyzing the sequence of Olambda to see of the origin has been completely knocked out. After looking at last year's files and the OriT sequence, we've determined that they have stuck the cassette into only part of the OriT sequence, which should have disrupted it but we are not positive if the left over region still has activity.
We did a genomic purification of wild type F plasmid and Olambda, and are performing a mapping of a PCR product of both using two different sets of oligos, OriTForward/OriTbackwardBB (which determines if the full length OriT is present) and ca941R/OriTbackwardBB (which shows if the KO cassette is at OriT). We expect Olambda to come back negative for OriTForward and positive for CA941R and the wild type to be positve for OriTForward and negative for OriTbackwardBB.
Lastly, more TriA and TriK plates were poured.
--Samanthaliang 14:04, 5 July 2006 (EDT)
SL_07/03/06
Last Friday, we discovered that pSB1A2-J01001 was not the plasmid we wanted to use for the Rlambda experiments as the TraJ carrier. Instead, the plasmid we want is pSB1A2-J01003. Also, the sequences for 1064, 1024, 1001, and 1093 all came back positive, but we are waiting for the sequence for J01003 in order to confirm its identity before we proceed with experiments.
As for the restreaking experiment with J01003 on Carb, Kan, and Cam plates that Will did last Friday from the -80 stocks, colonies only grew on the Carb plate, as expected.
Today, we're making chemically competent cells of BW20767 (the equivalent of Olambda for the R plasmid) but on a much smaller scale than the others (50mL rather than 500mL) because we wont need much of these cells.
After making these competent cells, we will transform J01003 into BW20767 by heat shock this time, because last time the electro-competent cells that we transformed J01003 into did not grow any colonies.
We're also performing the F mating experiment today again with Olambda, in order to see the neccessity of OriT for plasmid transfer during conjugaton. We are going to mate the following cells:
- Olambda with EC100D::pir+ (plate on TriK)
- pSB1A2-J01064 from -80 with EC100D::pir+ (plate on TriA)
- pSB1A2-J01064 into Olambda with EC100D::pir+ (plate on TriK)
- pSB1A2-J01064 Olambda with EC100D::pir+ (plate on TriA)
--Samanthaliang 18:27, 3 July 2006 (EDT)
JL_06/30/06
Since our results from the OriT/TraJ experiments were not as we expected, we will wait until we get the sequence results of J01064, J01093, J01001, and J01024 before doing any further experiments. Also, instead of using J01001, we will now be using J01003. Therefore, we sent J01003 out for sequencing today. Today, we also talked and shared information about our experiments and learned about the Ribosome team's experiments.
MF_06/29/06
Put colonies of BW20267 and Olam into 50ul of LB to grow them up. Olam grew quickly but BW20267 seems to be a slow grower. I also made TSS solution so Jen/Sam can make KCM competent cells. Grew up EC100D (tri resistant) to mix with the Spec resistant cells to see if the combined cells retained Spec resistance.
Results from 6/28/06
It turns out PSB1A2-J01001 is Kan and Amp resistant and we wanted to demonstrate only Amp resistance. So, we are going to transform JO1001 into new plasmid (PSB1A3) to get rid of Kan resistance.
Also, the sequences came back for clones 1 and 2 of pSB1A2-J23003 and they both look like matches, particularly for the double terminator.
--Mfleming 17:17, 29 June 2006 (EDT)
JL_06/28/06
Results from 6/27/06 Kan, Amp, Tri resistence experiment
Note: * means that the result does not match our hypotheses
| Culture | K | A | C | Tri | K results | A results | Tri results |
|---|---|---|---|---|---|---|---|
| 1 | TlamOlam | 1064 | - | - | Y(purple) | Y(purple) | N |
| 2 | TlamOlam | - | 1093 | - | Y(white) | N | N |
| 3 | TlamOlam | 1064 | 1093 | - | Y(white) | Y(white) | N |
| 4 | - | 1064 | - | - | N | Y(purple) | N |
| 5 | Fplasmid | - | - | - | Y(white) | N | N |
| 6 | Rlam | - | 1024 | - | Y(white) | N | N |
| 7 | Rlam | 1001 | 1024 | - | Y(pink) | Y(pink) | N |
| 8 | Rlam | 1001 | - | - | Y(white) | Y(white) | N |
| 9 | - | 1001 | - | - | Y*(white) | Y(white) | N |
| 10 | RP4 | - | - | - | Y(white) | Y*(white) | N |
| 11 | - | - | - | EC100D:pir+ | N | N | Y(white) |
Results from 6/27/06 OriT/TraJ Experiment (with pBad Promoter for TlambdaOlambda experiments)
| Culture | OriT | TraJ | # of Colonies on TriK | # of Colonies on TriA | Comments |
|---|---|---|---|---|---|
| 1 | + | - | 50 | >1000 | weird |
| 2 | - | + | 50 | n/a | |
| 3 | + | + | 70 | 8 | |
| 4 | + | - | n/a | 100 | |
| 5 | + | + | >1000 | n/a | looks good |
| 6 | - | + | 15 | 0 | looks good |
| 7 | + | + | 30 | >1000 | |
| 8 | + | - | 5 | n/a | looks good |
| 9 | + | - | n/a | >1000 | |
| 10 | + | + | >1000 | n/a | looks good |
Since our results from the "6/27/06 Kan, Amp, Tri resistence experiment" showed that J01001 was Kan and Amp resistent (and we believe it should only be Amp resistent) we are doing an experiment to see if J01001 is really KanR. We transformed J01001 into TGI cells and plated them on Amp and on Kan.
Also, our results from today's and yesterday's OriT/TraJ Experiments were unclear, so we are going to eliminate a variable by using Olambda (which doesn't have the TraJ knock out) instead of TlambdaOlambda. Today, we replated Olambda from the -80 stocks on three different plates: LB only, Kan, and Cam. We also plated the R plasmid equivalent of Olambda, BW207G7 on LB only because it doesn't have an antibiotic marker. We're growing these cells to competence in order to transform pSB1A2-J01064 into Olambda, and pSB1A2-J01001 into BW207G7 to test for color and transfer and then we are going to mate them with EC100D::pir+ (all this will hopefully be done tomorrow).
Finally, the plate with from the marker swap experiment with Rlambda and RP4-G looked good today with pickable colonies, so we spotted them and tested colonies for Spec Resistance and Kan Sensitivity. Eventually we will also test for Tet and Amp resistance.
--Jenlu 17:00, 28 June 2006 (EDT)
SIL_06/27/06
Results from 6/26/06 OriT/TraJ Experiment
| Culture | OriT | TraJ | # of Colonies on TriK | # of Colonies on TriA | Comments |
|---|---|---|---|---|---|
| 1 | + | - | 6 | 2 | looks good |
| 2 | - | + | 7 | n/a | missing pBad promoter |
| 3 | + | + | 3 | 5 | missing pBad promoter |
| 4 | + | - | n/a | 4 | looks good |
| 5 | + | + | >1000 | n/a | looks good |
| 6 | - | + | 6 | 200 | wierd, hoping for a smaller TriA count, but possible |
| 7 | + | + | 3 | 1 | possible tube switch with 9 |
| 8 | + | - | 0 | n/a | looks good |
| 9 | + | - | n/a | >1000 | possible tube switch with 7 |
| 10 | + | + | >1000 | n/a | looks good |
Today we redid the experiment, using Arg. with the F plasmids for the Pbad promoter, and the R plasmids the same as before, in order to see if tube 7 and 9 really were switched.
The diluted restreaking of Rlambda and RP4-6 from yesterday failed and a lawn was still achieved, so we redid that again this time, adding another 10 fold dilution. We also poured more TriA and TriK plates.
Also we streaked all of the plasmid combinations (1-10) from the OriT and TraJ experiments onto a Kan plate, an Amp plate, and a Tri plate, in order to make sure that all the plasmids have the antibiotic resistances that we think they have, and that they don't have any extras.
--Samanthaliang 14:41, 27 June 2006 (EDT)
SIL_06/26/06
Today we did the TraJF/oriTF experiment. We Conjugated EC100D:pr+ with:
- TlambdaOlambda+1064 (plated on TriA and TriK)
- TlambdaOlambda+1093 (plated on TriK)
- TlambdaOlambda+1064+1093 (plated on TriA and TriK)
- DH10B/1064 from -80 (plated on TriA)
- F plasmid on Kan
(Plated on TriK) - Rlambda+1024 (Plated on TriA and TriK)
- Rlambda+1024/1001 (Plated on TriA and TriK)
- Rlamdbda+1001 (Plated on TriK)
- DH10B/1001 from -80 (Plated on TriA)
- wt RP4 (Plated on TriK)
Also, we should note, that the TlambdaOlambda+1064 and the DH10B/1064 cells were the only ones that were really red. Rlambda+1024 was a peachy color. TlamdbdaOlambda+1064_1093 and Rlambda+1024/1001 were both a very very faint shade of pink, which is troublesome, since they should have come out red as well.
We also mini-prepped 4 clones of pSB1A2-J23003(TraJr-TT)and created an APE file for it. We had to also sequence them because mapping was not possible because the distinguishing fragment from pSB1A2-J01001, which J23003 was constructed from, the terminator was too small of a sequence and no enzymes could be used to cut so that there was an obvious difference between the two plasmids when mapped.
Lastly, the conjugation experiment with Rlambda and RP4-G that we did on Friday worked very well. In fact, it was too efficient and ended up as a dense lawn. Thus, we restreaked the plate, but only after diluting it from 1uL to 1mL LB twice, then taking only 24uL of the diluted cell solution and plating it.
--Samanthaliang 17:08, 26 June 2006 (EDT)
JL&SIL_06/22/06
Today we mapped pSB3C6-J01024 using EcoRI and SpeI and pSB3C6-J01093 BamHI and SpeI. We had 100% recovery of 4 clones of each, and did not get any colonies of just the backbone alone. This was determined by a analytic digestion which resulted in 2 stripes for each of the 8 columns we ran in the gel (if it had only been 1 stripe, that would've indicated the colony was that of the the backbone pAC997). Because of these positive results, I made -80 stocks of the clone 1s of both pSB3C6-J01024 and pSB3C6-J01093, and put the minipreps of clone 1 and 2 of each in the freezer.
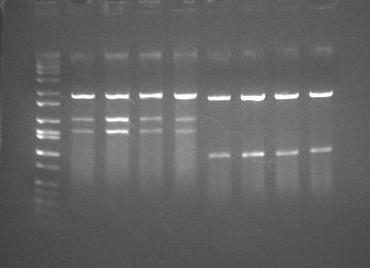 (ladder, 4 clones of J01024, 4 clones of J01093)
(ladder, 4 clones of J01024, 4 clones of J01093)
We also digested, gel purified, ligased J01001 and B0015, and transformed by heat shock to get the product J23003 (TraJf-tt).
We also replated RP4 and F on KAN plates, EC100D:pir+ on TRI plates, and 1064 and 1001 on AMP plates. Then we picked colonies from the Rlambda plate from 6/19/06 to grow into competent cells.
Lastly, we are preparing for a big experiment regarding the TlambdaOlambda cells, by transforming plasmid pSB1A2-J01064 into one sample, plasmid pSB3C6-J01093 into another sample, and both of those plasmids into a third sample (this is the one we are most concerned about) using heat shock.
The purpose of this experiment would be to determine whether or not the F plasmid with the KAN marker, J01064 with the AMP marker and OriT, and J01093 with a CAM marker and TraJf will transform into a new plasmid with a TRI marker.
--Samanthaliang 20:11, 22 June 2006 (EDT)
SIL_06/19/06
Today we subcloned pSB1A2-J01024 into the pAC997 backbone to create the final product pSB3C6-J01024. We did this by digesting pSB1A2-J01024, gel purifying it, ligating it, and then heat shocking it. We also subcloned pSB1A2-J01093 into the pAC997 bacbone to create pSB3C6-J01093, starting from a PCR product of J01093 because in a previous experiment J01093's results came out unclean. This was also done through digestion, gel purification, ligation and heat shock.
We also restreaked Rlambda from the -80 freezer stocks onto a Kan plate.
Lastly, we made sure that J01001 was in the freezer for future use.
--Samanthaliang 19:17, 19 June 2006 (EDT)
JCA_06/17/06
We had some difficulty with the traJF and traJR stocks for our initial tests of conjugation. It is unclear what plasmid the -80 stocks are inserted into. We tried to grow up the "J01024" from Box2>7v4 in carbencillin media. It didn't grow. Yesterday we grew up the "J01024/KAN" in Box1>9v1. It grew just fine and the cell pellet was slightly pink due to its Ptet-RFP cassette. It is unclear whether this is pSB1AK3 or perhaps a pSB*K* plasmid? Unclear, but we're going to transfer it into pSB3C6 anyway, so who cares. In addition, I was concerned that the oriTF plasmids might have CmR markers on them. The biobricks annotation for the J01002 from the plates indicated it was pSB1AC3. That would be bad--it needs to be just ampR. So, we grew up the two different J01002's from the freezer stocks (Box1>6v4) and (Box1>8v2). The second one for some reason is not in the slot indicated on the -80 stocks page. Someone will need to re-update the -80 stocks page. Dan spotted the two J01002 strains and J01064 from Box1>5v1 (oriTF/OnRFP). All three grew spots on amp but not on Cm or Kan. So, they are all 3 on pSB1A* plasmids, so it's all good. Incidently, J01064 was flaming pink in cell pellets. Much brighter than J01024 despite them having the same RFP cassette. I miniprepped J01024 and J01064.
JL_06/07/06
Goal: We want to determine the antibiotics resistence of the F and R knockout plasmids (TlambdaOlambda)
Results:
We grew TlambdaOlambdaRlambda to early log density. We then streaked the cultures with pipette tips on LB agar plates containing the following anti-biotics:
- 15ug/mL gentamicin (G)
- 50ug/mL ampicillin (A)
- 25ug/mL chloramphenical (C) ---errr, axe that datapoint, the plate disapeared.--JCAnderson 13:45, 8 June 2006 (EDT)
- 50ug/mL spectinamycin (Sp)
- 25ug/mL tetracycline (Tet)
- 20ug/mL trimethylprim (Trim)
TlambdaOlambda grerw on kan only
Rlambda grew on kan and tet only--JCAnderson 19:07, 7 June 2006 (EDT)