Beauchamp:ProcessDiffTensImgData
These steps show how to process DTI data acquired on the UT Philips 3T scanner
Create a 3d+time DWI BRIK from DICOM files
User Note: Press "T" in DTI Query to remove the VOI label
1) The first step is to reconstruct the raw diffusion-weighted volumes ...
Set the subject identifier variable. This is just an example: you can use any string.
set ec="TA309"
Make the .BRIK and .HEAD files from raw data stored in the subdirectory {$ec}_060807 (your directory name will be different)
You need to know number of dynamics (34), number of slices (70), and repetition time (9543.7 ms)
The dynamics, slices and repetition time are all available in the Philips .PAR file
The output is an AFNI image file called {$ec}_DMaps+orig
to3d -session . -epan -prefix {$ec}_DMaps -time:tz 34 70 9543.7 alt+z ../{$ec}_060807/ax_dti_high/DICOM/IM_* ../{$ec}_060807/ax_dti_high/DICOM/00000001/IM_*
Correct DWI for Movement and Align Volumes to High Resolution MRI
2) The second step is to correct diffusion volumes for movement, and coregister the data to the space of the high resolution anatomical volume
This step registers each diffusion weighted volume to the skull-stripped anatomical volume, in this case called {$ec}avaganatSS+orig
The output is an AFNI image file called {$ec}DWIREGtoSSanat and a textfile containing the transformation parameters called {$ec}DWIREGtoSSanatXform
3dAllineate -base {$ec}avganatSS+orig -source {$ec}_DMaps+orig -prefix {$ec}DWIREGtoSSanat -verb -warp affine_general -EPI -cost mutualinfo -1Dfile {$ec}DWIREGtoSSanatXform -master {$ec}_DMaps+orig
Compute the Diffusion Tensor in Scanner Space
3) The third step is to compute the diffusion tensor image, which will be used for deterministic fiber tracking
This AFNI program 3dDWItoDT computes the tensor from the diffusion volumes acquired with different gradient orientations
The gradient orientation table is stored in a textfile called DWGradientVectors.txt, and is listed below the tensor computation command
The gradient orientations are specific to the Philips 32 direction diffusion encoding scheme (high angular resolution) with the gradient overplus option
The output is a file called {$ec}_Tensors.nii.gz, which is in single-subject space and in alignment with the high resolution anatomy
3dDWItoDT -prefix {$ec}_Tensors.nii.gz -automask -reweight -max_iter 10 -max_iter_rw 10 DWGradientVectors.txt {$ec}DWIREGtoSSanat+orig'[0..32]
The x,y,z gradient orientations (contents of DWGradientVectors.txt)
-0.532, -0.465, -0.707
-0.532, -0.465, 0.707
0.658, -0.753, 0.000
-0.670, -0.226, -0.707
-0.254, -0.660, -0.707
-0.028, -0.707, -0.707
0.382, -0.595, -0.707
0.678, -0.201, -0.707
-0.670, -0.226, -0.707
-0.340, -0.686, -0.644
0.246, -0.725, -0.644
0.481, -0.520, -0.706
0.686, -0.340, -0.644
-0.737, -0.424, -0.526
-0.519, -0.674, -0.526
0.511, -0.680, -0.526
0.674, -0.519, -0.526
-0.753, -0.658, 0.000
-0.737, -0.424, 0.526
0.674, -0.519, 0.526
0.424, -0.737, 0.526
0.674, -0.519, 0.526
-0.664, -0.382, 0.644
-0.752, -0.657, 0.055
-0.340, -0.686, 0.644
0.245, -0.725, 0.644
0.658, -0.753, 0.008
0.686, -0.340, 0.644
-0.752, -0.657, 0.049
-0.013, -0.707, 0.707
0.658, -0.753, 0.012
0.706, -0.047, 0.707
Talairach Transform the Aligned Volumes
4) If you've transformed your skull-stripped anatomical volume to Talairach or MNI space, you may apply this transform to the individual diffusion-weighted volumes
This allows you to conduct fiber tracking in a standardized space to allow for comparisons between subjects.
To do this use the @auto_talairach script which is distributed with AFNI
The output is a BRIK of all the diffusion weighted volumes each in Talairach space with 2mm isotropic resolution
@auto_tlrc -apar {$ec}avganatSS_at+tlrc. -input {$ec}DWIREGtoSSanat+orig. -dxyz 2 -verb
Compute the Diffusion Tensor in Talairach Space
5) Now we compute the tensor on the Talairach transformed diffusion volumes
The output is a tensor file similar to the file produced in step 3, except that it is in standard Talairach space not single-subject space
3dDWItoDT -prefix {$ec}_Tensors_at.nii.gz -automask -reweight -max_iter 10 -max_iter_rw 10 DWGradientVectors.txt {$ec}DWIREGtoSSanat_at+tlrc'[0..32]
Compute FA Volumes and Whole-Brain Fiber Pathways
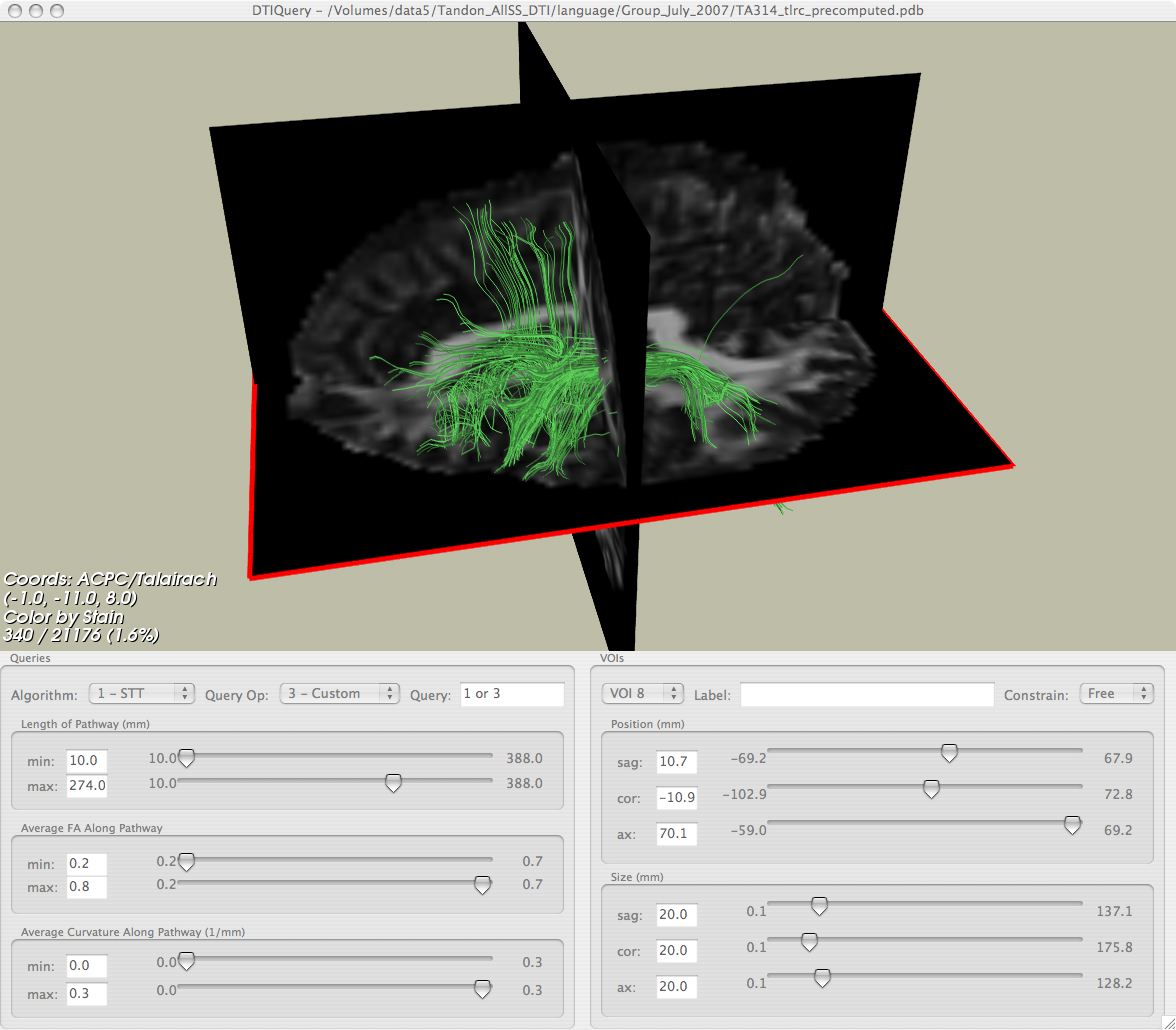
6) The last step is to compute fractional anisotropy (FA) volumes and whole-brain fiber pathways with DTI Query v1.1b
DTI Query 1.1b is available at: http://graphics.stanford.edu/projects/dti/dti-query/
The command below presumes you have installed DTI Query in your /Applications directory
First we do computations on the data in single-subject space
The output files are fa.nii.gz and precomputed.pbd, which are the FA volume and fiber pathways respectively
They are renamed here to have the subject identifier pre-pended and the label orig
/Applications/DTIQuery_v1.1macintel/dtiprecompute_cmd -t {$ec}_Tensors.nii.gz -o . -f 0.15 -a 45 -d 2.0 -s 4.0 -minL 5.0 -maxL 300.0 -i EULER
mv fa.nii.gz {$ec}_orig_fa.nii.gz
mv precomputed.pdb {$ec}_orig_precomputed.pdb
Next we do computations on the data in standardized Talairach space
The output files are fa.nii.gz and precomputed.pbd, which are the FA volume and fiber pathways respectively
They are renamed here to have the subject identifier pre-pended and the label tlrc
/Applications/DTIQuery_v1.1macintel/dtiprecompute_cmd -t {$ec}_Tensors_at.nii.gz -o . -f 0.15 -a 45 -d 2.0 -s 4.0 -minL 5.0 -maxL 300.0 -i EULER
mv fa.nii.gz {$ec}_tlrc_fa.nii.gz
mv precomputed.pdb {$ec}_tlrc_precomputed.pdb
Interactively Visualize Fiber Pathways
Now you are ready to load your processed FA and fiber pathway file in DTI Query for interactive tractography!
Notes on Eddy Current Correction
To reduce Eddy Currents, Phillips recommends increasing the partial k-space collected from 0.5 to 0.73; Vips does this as a matter of course. Phillips does not support the double spin-echo method described below, but YuXiang has implemented it and finds that it gives very similar results to the standard Phillips sequence. Note the importance of 12-parameter registration with 3dAllineate:
- here, alignment of each DWI to skull stripped anatomy is done with the warp affine_general (12 parameter option)
3dAllineate -base {$ec}anatSS+orig -source {$ec}_DMaps+orig -prefix {$ec}DWIREGtoSSanat -verb -warp affine_general -EPI -cost mutualinfo -1Dfile {$ec}DWIREGtoSSanatXform -master {$ec}_DMaps+orig
- compute the diffusion tensors; Daniel Glen says the –reweight option should provide additional help to reduce eddy current distortion
3dDWItoDT -prefix {$ec}_Tensors -automask -reweight -max_iter 10 -max_iter_rw 10 DWGradientVectorsAfter12-21-07.txt {$ec}DWIREGtoSSanat+orig'[0..32]'
Eddy-currents can be a problem, but there aren't that many reliable
correction methods out there. The FSL method is just a matter of
performing affine registration to the b=0 image, which isn't
necessarily the best approach given that the b=0 image has a large CSF
contribution, which is absent from the DW images - in certain cases,
that's been shown to introduce 'over-stretching' of the DW images to
fit the extra ring of CSF. That shouldn't however be a problem with
your data, since they seem to have very good SNR.
In general, the most effective approach is to use the so-called 'twice-refocused' acquisition, which is designed to minimise eddy-current effects - it is effectively a double spin-echo sequence. Here's the reference if you need it:
Reese TG, Heid O, Weisskoff RM, Wedeen VJ Reduction of eddy-current-induced distortion in diffusion MRI using a twice-refocused spin echo Magn Reson Med 2003; 49 (1): 177-182
It is the default of Siemens systems, and has been for several years now. I can't tell from the DICOM data you sent me whether your data were acquired with a single or double spin-echo sequence, but I would be surprised if Philips didn't also at least offer that sequence. If your data were acquired with the twice-refocused sequence, then distortions due to eddy-currents should be negligible, or at least sub-voxel. If they were acquired with a single SE sequence, then you do unfortunately need to correct for eddy-currents.
However, what I noticed was that the DICOM data you sent me had a fair bit of motion artefact, which had obviously been corrected in the equivalent NIfTI image. Whatever routine you used to do the motion correction did a very good job, and the resulting images do not seem to have any significant eddy-current artefact in them - you can usually tell by looking at the colour-coded direction maps: with eddy-current artefacts, there's usually a ring of colour around the edges of the brain, where the various DW images don't match. It's possible that your motion correction also performs eddy-current correction as a by-product. This will be the case in particular if you used a full affine registration algorithm (i.e. 12 degrees of freedom, including scaling & shearing), rather than a simpler rigid-body algorithm (i.e. 6 degrees of freedom, only translation & rotation). For example, the eddy-correct routine in FSL uses such an affine registration routine, which explicitly allows scaling, shearing & translation (all possible consequences of eddy-current effects). You might want to check what algorithm you used for the motion correction, and in particular what parameters were used, as you may already be performing eddy-current correction...
Hope that helps,
Donald.
Scripts For Processing DTI Data
File:@preprocess fsl.csh File:Gradients.zip
It can also be good to modify the gradient matrix to take into account subject movement. (Since gradients are applied in scanner space, not subject space).
A compelling paper on this topic is Leemans and Jones, Magnetic Resonance in Medicine, 2009
Here is Tim's script to do that. File:@rotgraddirs.csh