Beauchamp:AnalyzeEEGinMatlab
Data from the Nihon Kohden EEG acquisition systems are stored in a proprietary format
To analyze these data in a flexible analysis environment like Matlab, you must first export them as an ASCII textfile from the Nihon Kohden Review program.
The Review program may only be installed on a PC running Windows XP.
This Wiki page shows how to install the Nihon Khoden review program, read EEG data into it, export data to ASCII, and convert ASCII files to matlab files.
Installing the Nihon Kohden Software
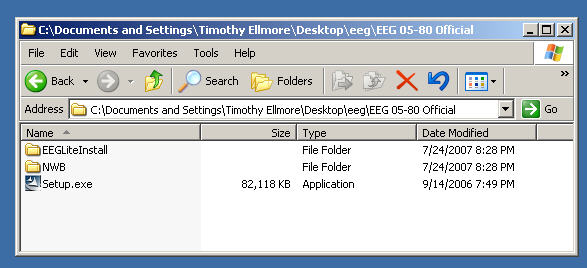
There is a self-extracting Windows executable file on data8 called "EEG 05-80 11.06 Official Release.exe"
Double click on this executable to extract files onto your desktop. A directory will be created called "EEG 05-80 Official" with these contents:
Placing Raw EEG Data into the Appropriate Directory
Double click on the Setup.exe to install the Nihon Kohden software in C:\
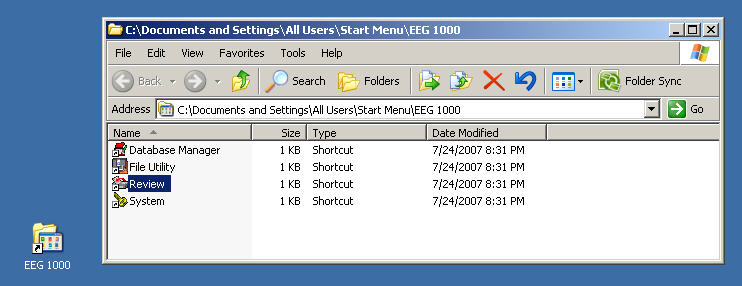
A directory called "EEG 1000" will be placed on your Windows desktop that contains four executables.
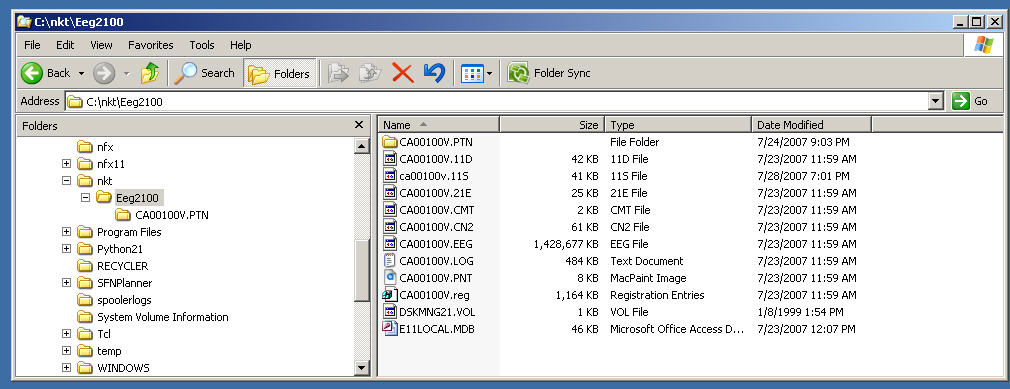
A directory called C:\nkt\Eeg2011 will also be created.
You must copy the raw Nihon Kohden EEG data from a particular session into this directory.
The Review progam automatically looks in this directory and will read whichever session's data is in there. Here's an example of what this directory may contain after you've copied the raw files:
Viewing Raw EEG Data in the Review Program
Now run the Review program from the EEG 1000 folder on your desktop.
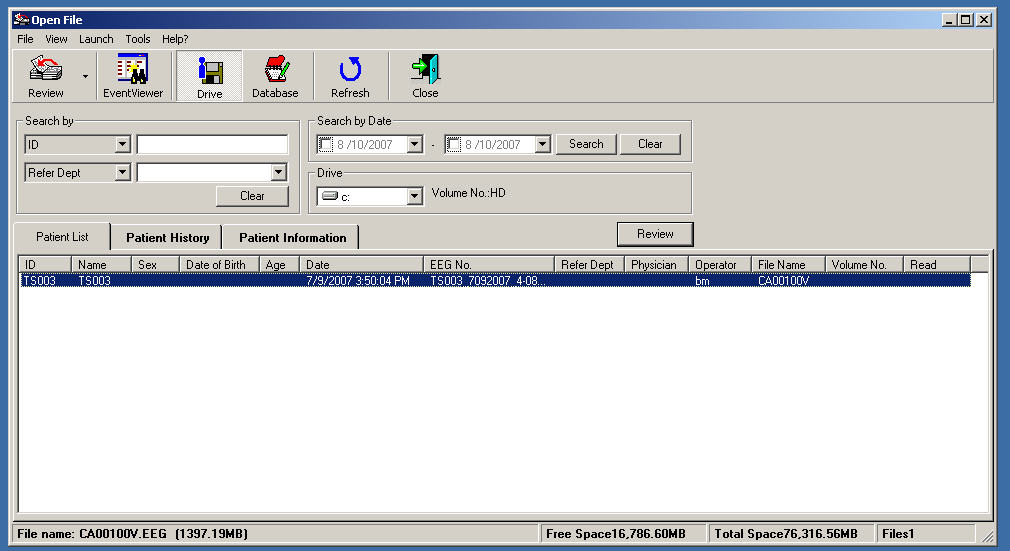
You should see information about the particular EEG session data you placed in C:\nkt\Eeg2011 in a dialog box that looks like this:
Choosing EEG Data Time Segments for Export
Click on subject ID field, and then click on the Review button in the upper left corner and you should see the EEG data displayed in the Review window
Now you are ready to identify a segment of EEG data and export it as an ASCII file.
If the EEG tech has made annotations about when a particular task or set of event occured you will see them listed in a box on the right.
Choose a given event marker to start at that time, then choose "Wave Select" from the "Tool" menu.
Advance to the time your event or task has ended and choose "Set" in the "Wave Select" box.
In the example below the data from 16:29 to 16:44 will be extracted.
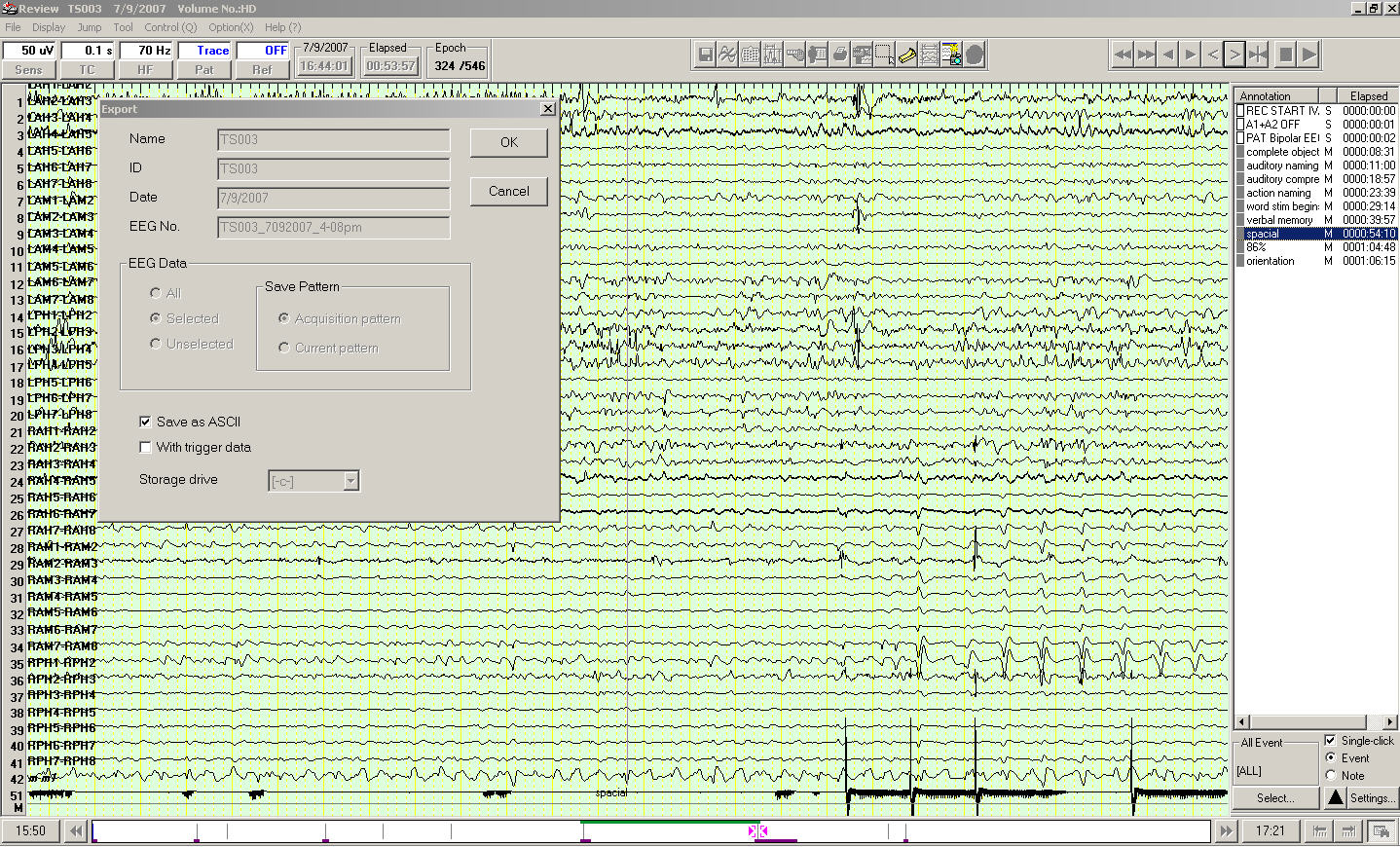
Exporting EEG Data as ASCII Files
Click the "Set" button and you will see the "Export" box. Choose "Save as ASCII" and click "OK"
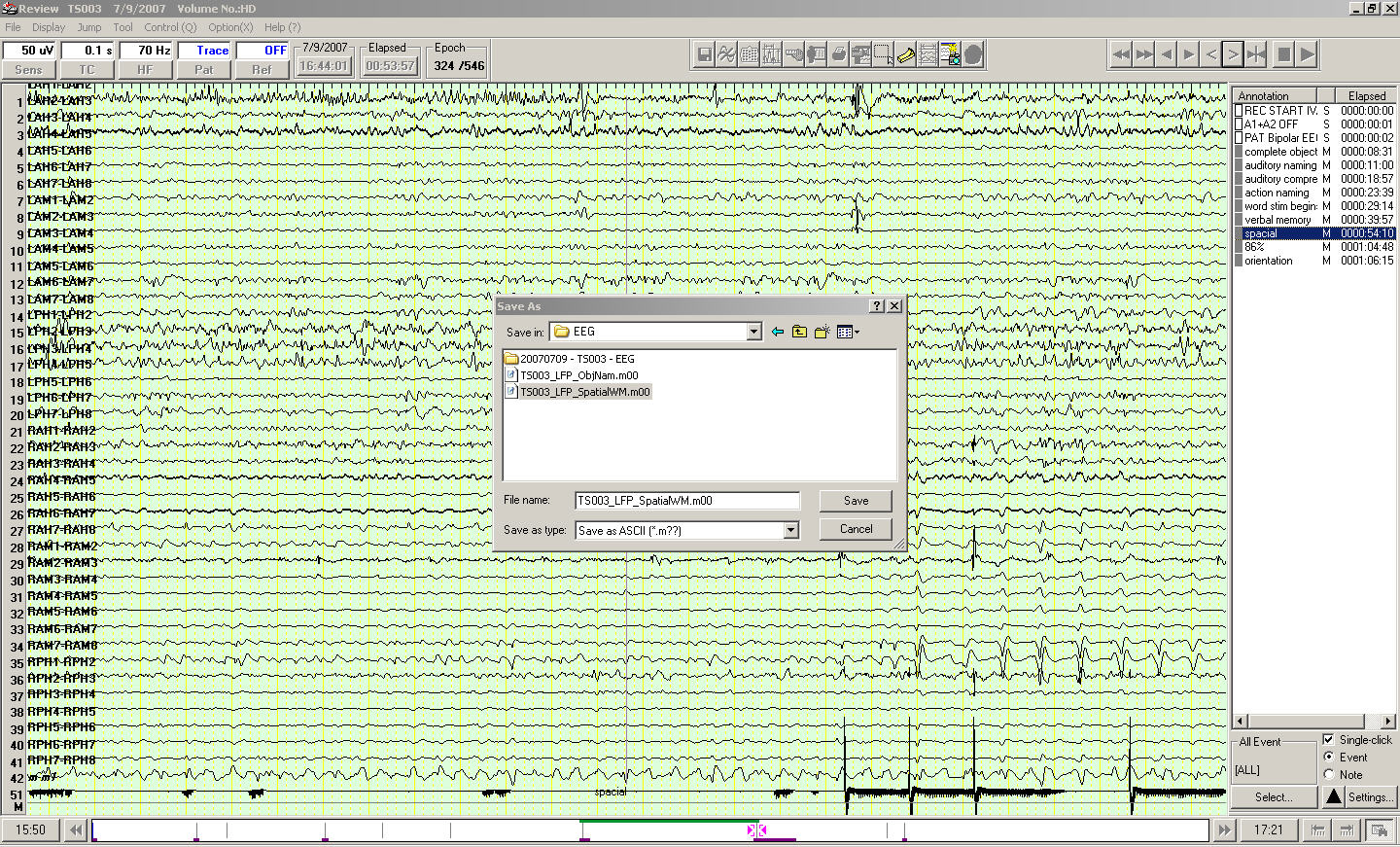
Finally, choose a directory and type a name for the output ASCII textfile. Export may take a while depending on the EEG segment length and sampling rate.
Converting the ASCII output to a Matlab file
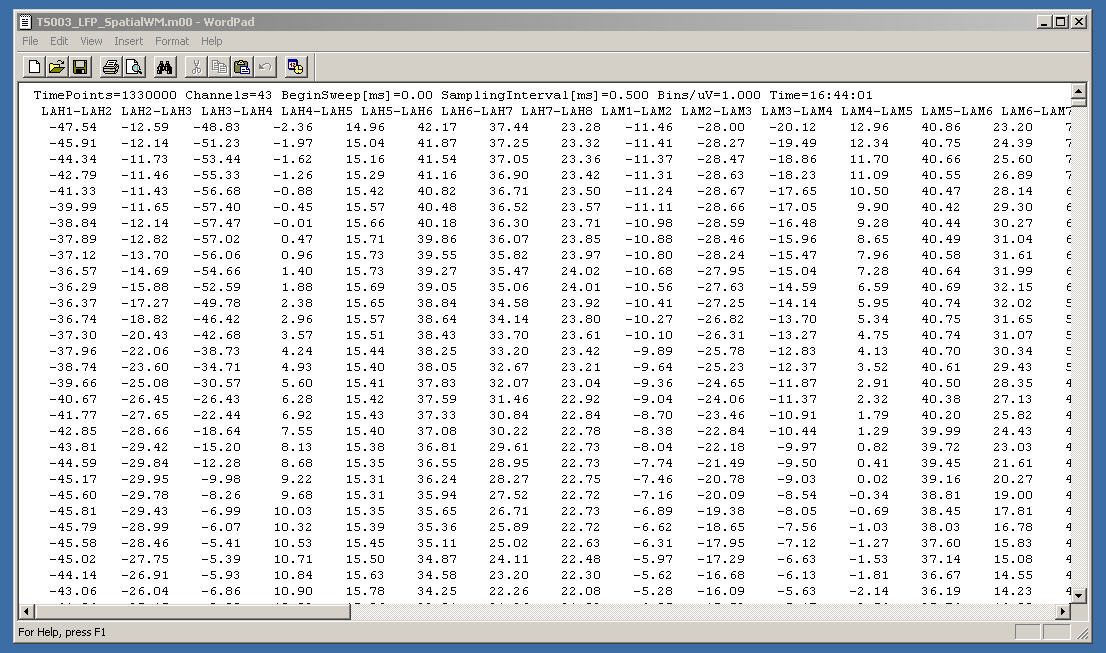
If you were to open the output ASCII file in a text editor (not recommended because they can be enormous), you would see something like this:
The first two lines contain information about how the data were sampled and the names of the recorded channels.
The next lines contain the data with each channel listed by column and each row is a sampled data point.
These ASCII textfiles can easily exceed 1GB (e.g., 11 mins of 43 channels sampled at 2000 Hz ~ 500 MB).
Economy of storage may be achieved by storing these data in a single matlab structure. The eeg data is stored as a 16-bit short integer matrix in one of the structure's fields.
The matlab code to convert a Nihon Kohden ASCII file is provided by a function called convert_nkascii2mat.m:
function convert_nkascii2mat(fname,matfname,mult)
% Reads a Nihon Kohden EEG file in ASCII format (fname), converts
% the floating point ASCII values to 16-bit signed integers, and writes
% them to disk as a matlab structure file (matfname).
%
% The mat file holds a structure nkdata with the following fields:
% nkdata.eeg - all the eeg data stored as nchannels-by-npoints matrix
% nkdata.multiplier - a multiplier; divide stored value by this to floats
% nkdata.num_dpoints - number of data points per channel
% nkdata.nchannels - number of recorded channels
% nkdata.bsweep - begin sweep (ms)
% nkdata.ms_per_sample - ms per data sample
% nkdata.binsuV - bins per microvolt
% nkdata.start_time - start time of exported data segment
% nkdata.ch_names - char array of channel names
%
% Example:
% convert_nkascii2mat('TS003_LFP_SpatialWM.m00','TS003_LFP_SpatialWM',10);
%
% Note:
% Data in the NK ASCII file are stored with floating point precision to the hundreth
% decimal place. On conversion, each data value is multiplied by 10 in order to store
% data as 16 bit integers (storage as 16 bit int rather than 32 bit float saves space).
% Therfore, when reading the resultant mat file each data value must be divided by the
% nkdata.multiplier, which is given by mult, the 3rd command line parameter.
% You must do this to preserve micovolt units. Here's how:
%
% load TS003_LFP_SpatialWM.mat
% nkdata.eeg=nkdata.eeg./nkdata.multiplier;
%
% Last modified 8/10/07 by TME
% Help: Timothy.Ellmore@uth.tmc.edu
% get header information
[tpoints,nchannels,bsweep,sampintms,binsuV,start_time,ch_names]=get_nkheader(fname);
% the name of the ascii file
fid=fopen(fname,'r');
% skip header information
hline1=fgets(fid);
hline2=fgets(fid);
% read file in 10 separate chunks (helps save RAM)
nsegs=10;
seg_len=tpoints/nsegs;
fprintf('\nWill read %d segments each with %d sampled points\n',nsegs,seg_len);
nkdata.eeg=[];
lastp=1;
for seg_num=1:nsegs
dc=[];df=[];dtmp=[];
if(seg_num==1)
dc=textscan(fid,'%f',nchannels*seg_len,'HeaderLines',0);
else
dc=textscan(fid,'%f',nchannels*seg_len,'HeaderLines',0);
end
df=dc{1};
df=df.*mult; % multiply by 10, don't forget to divide when reading in data!
fprintf('\n... converting ASCII floats to 16-bit signed ints ...',seg_num*nsegs);
npoints=length(df)/nchannels;
dtmp=reshape(df,nchannels,npoints);
clear df dc;
% store in cumulative matrix
nkdata.eeg(1:nchannels,lastp:(lastp+npoints-1))=dtmp;
nkdata.eeg=int16(nkdata.eeg);
clear dtmp df dc;
fprintf('\n%d to %d, %d percent completed!\n',lastp,lastp+npoints-1,seg_num*nsegs);
lastp=lastp+npoints;
end
fclose(fid);
% store other information in nkdata structure
nkdata.multiplier=mult; % scale
% individual pieces of information from header line 1
[nkdata.num_dpoints,nkdata.nchannels,nkdata.bsweep,nkdata.ms_per_sample,nkdata.binsuV,nkdata.start_time,ch_names]=get_nkheader(fname);
% sampling rate in Hz
nkdata.sampHz=1000./nkdata.ms_per_sample;
% channel names
nkdata.ch_names=ch_names;
% write a mat file to disk
eval(sprintf('save %s nkdata',matfname));
convert_nkascii2mat.m uses another function to parse the two lines of header information in the ASCII file. This function is called: get_nkheader.m:
function [ntpoints,nchannels,bsweep,sampintms,binsuV,start_time,ch_names]=get_nkheader(fname)
% Reads a Nihon Kohden EEG file in ASCII format (fname) and
% returns 7 pieces of information from the header
%
% The function returns the following information:
% ntpoints - number of data points per channel
% nchannels - number of channels sampled during recording
% bsweep - begin sweep (ms)
% sampintms - sampling interval in ms
% binsuV - number of bins per microvolts
% start_time - starting time of recording
% ch_names - char array of channel names
%
% Example:
% [tpoints,nchannels,bsweep,sampintms,binsuV,start_time,ch_names]=get_nkheader('TS003_LFP_SpatialWM.m00');
%
% Last modified 8/19/07 by TME
% Help: Timothy.Ellmore@uth.tmc.edu
% the name of the ascii file
fid=fopen(fname,'r');
%header line 1: acquisition information
hline1 = fgets(fid);
% get the six different fields in the header, store in cell array
hd=textscan(hline1,'%s %s %s %s %s %s');
fprintf('\nNihon Kohden ASCII EEG header fields:');
fprintf('\n-------------------------------------');
% number of timepoints
[txt,ntpoints]=strread(char(hd{1}),'%s%d','delimiter','=');
fprintf('\n%s is %d',char(txt),ntpoints);
% number of channels sampled
[txt,nchannels]=strread(char(hd{2}),'%s%d','delimiter','=');
fprintf('\nNumber of %s is %d',char(txt),nchannels);
% begin sweep in ms
[txt,bsweep]=strread(char(hd{3}),'%s%f','delimiter','=');
fprintf('\n%s is %2.2f',char(txt),bsweep);
% sampling interval in ms
[txt,sampintms]=strread(char(hd{4}),'%s%f','delimiter','=');
fprintf('\n%s is %1.2f (or %2.1f Hz)',char(txt),sampintms,(1000./sampintms));
% bins per micro volt
[txt,binsuV]=strread(char(hd{5}),'%s%f','delimiter','=');
fprintf('\n%s is %1.2f',char(txt),binsuV);
% start time
tt=char(hd{6});
start_time=tt(end-7:end);
fprintf('\nStart Time is %s\n',start_time);
% header line 2: names of recording channels
hline2 = fgets(fid);
% channel names as cell array
ch_names=textscan(hline2,'%s');
% convert to char array
ch_names=char(ch_names{1});
% close input file
fclose(fid);
Using these matlab functions data can be converted from ASCII, loaded into matlab with no loss of precision, and plotted as shown in the following commands:
% convert the NK ASCII file, write matlab file
convert_nkascii2mat('TS003_LFP_SpatialWM.m00','TS003_LFP_SpatialWM');
% load the converted matlab file
load TS003_LFP_SpatialWM.mat;
% convert all eeg data values to microvolts
nkdata.eeg=nkdata.eeg./nkdata.multiplier;
% plot a single channel (in this case #2) vs. ms elapsed
plot(1:nkdata.ms_per_sample:((nkdata.num_dpoints+1)*nkdata.ms_per_sample),nkdata.eeg(2,:));
% show the name of recording channel #2
nkdata.ch_names(2,:)
--Stim 12:53, 21 August 2007 (CDT)