BME103:W930 Group8
| Home People Lab Write-Up 1 Lab Write-Up 2 Lab Write-Up 3 Course Logistics For Instructors Photos Wiki Editing Help | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Group 8 PCR Gurus
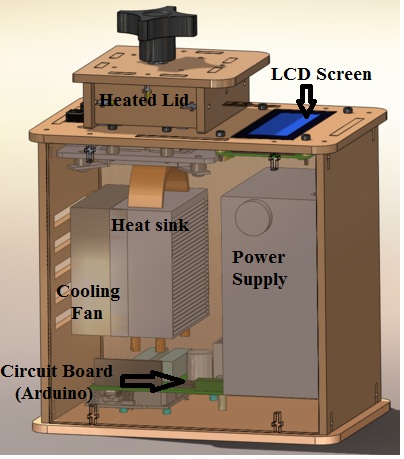
LAB 1 WRITE-UPInitial Machine TestingThe device depicted above is an open PCR(Polymerase Chain Reaction), and is used for the amplification of DNA. It works by heating and cooling the DNA samples allowing for the splitting and recombination of the DNA with the primers located in the solution. To accomplish this, the device connects to the USB port of a computer where the format of the cycles are inputted. From there the device repeatedly heats and cools via the heated lid, heat sink, and fan. The information is then fed to the Arduino board which controls the heating and cooling as well as transferring the information to the LCD screen. Experimenting With the Connections When we unplugged part the LCD screen(part 3) from the Arduino(part 6), the LCD display went black and ceased to work. When we unplugged the white wire that connects the Arduino(part 6) to the PCR block (part 2), the machine relayed a temperature reading of -40˚C, indicating a malfunction and the temperature could not be accurately determined.
ProtocolsPart 1: PCR Protocol
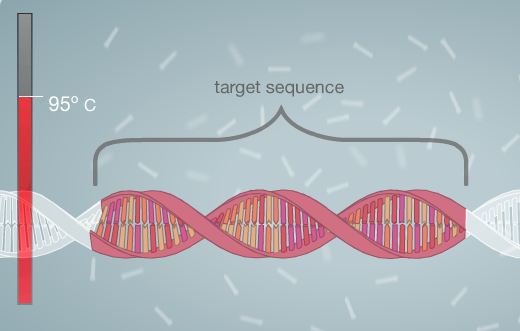
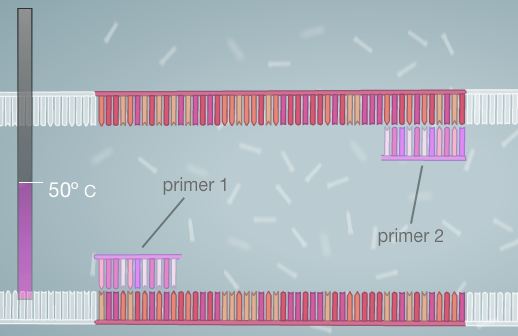
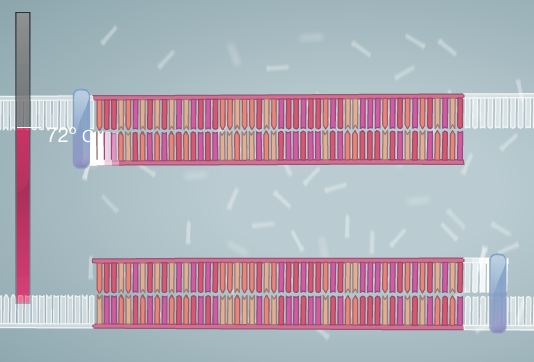
Polymerase Chain Reaction Polymerase Chain Reaction is used to amplify DNA. In order for this process to be successful, Template DNA must be replicated. A template of DNA consists of a strand in which the order of the nucleotides (bases) is known. With this information, it is possible to replicate the DNA strand by using heat, primers, and polymerase. The process begins by combining the DNA and the master mix. The master mix is composed of all of the necessary ingredients for the completion of the PCR process. The DNA is then placed into a PCR machine, or thermal cycler. The DNA is heated in the thermal cycler, allowing it to denature, or separate into two strands. The primers are then added to the template strand. They mark the start and end points of the specific sequence that is being targeted for replication. Next, the cycler is once again heated. This allows for the polymerase enzyme to activate and to begin adding nucleotides to the DNA strand. At the end of this process, two strands are created (essentially each strand of DNA has one side composed of new nucleotides that are added by the polymerase and the other composed of the original nucleotides, thus allowing for a total product of two strands). The machine is then cooled and re-heated again. This cycle is generally repeated for 20-30 cycles, or until the desired amount of target DNA sequence strands is reached. The composition of the master mix is listed below. Steps to Run PCR
Part 2: Flourimeter Protocol Flourimeter Measurements The flourimeter is used to measure the amount,or intensity of fluorescence. The blue LED light is turned on to begin using the device. A camera phone is placed onto the designated tray. In order for the process to work, the flash settings on the camera phone must be turned off. Also the ISO must be 800 or higher and exposure must be set to its maximum. It helps to turn off auto focus as well. A slide is placed onto the flourimeter. On this slide a drop of water is inserted in the middle of the first two rows using a pipette. Two more drops are added to the first drop. The LED light is aligned with the drop and the flourimeter is then covered using a light box. The light box will remove excess light, allowing for images of the drop of water to be taken. The drop is then removed from the slide.The process of putting drops into the slide is then repeated using the next set of holes on the slide (Move the slide so that the new set of holes is in line with the blue light before putting the next drop onto the slide. For both parts of the process, each drop should be between 130 and 160 micro-liters.) The process of taking photos of the drop is then repeated using the same method previously presented. About three photos should be taken for each drop.
Note: The computer will recognize that it is a picture file and save it as Jpeg, unless otherwise specified. Research and DevelopmentSpecific Cancer Marker Detection - The Underlying Technology This experiment uses PCR (polymerase chain reaction) to amplify DNA strands to detect certain base pair sequences. First, the Template DNA, DNA primers, salts (MgCl2 or Magnesium Chloride), Taq Polymerase and deoxynucleotide triphosphates (dNTPs) are added together into a tube. PCR starts with a heating that breaks apart the DNA bonding and creates two single strand DNA molecules. The process then cools and allows the primers to bind to the specific region on the single strand DNA. The Taq polymerase then takes the dNTPs (base pairs) and adds them onto the DNA single strand in the 3' direction of the primer. This creates two DNA strands out of the one that was first added into the solution. The heating and cooling process that occurs is called thermal cycling. This cycling is done 30-35 times to amplify the DNA to a point where it is easy to detect. The process of duplicating these DNA strands means that by the end of 30-35 cycles, there would be a clear amplification. Syber-Green dye was used to show double-stranded DNA as it fluoresces only when attached to double binded DNA. Our certain sequence rs17879961 is related to the susceptibility to Breast and Colorectal Cancer. This is a mutation in the CHEK2 gene. The CHEK2 gene is related to fixing DNA damage that occurs. These mutations cause the ability for DNA strands damaged to continue to create proteins when damaged. This creates a higher susceptibility to prostate and breast cancer. PCR would be able to find this mutation by using the primers shown below which are tailored for the rs17879961 mutation in the CHEK2 gene. This would allow for the test to predict susceptibility to certain cancers. The way PCR would do this would be that the primers would bind onto the mutation during the cooling process. This would then amplify the DNA as the primers would bind. If the primers didn't bind because the mutation didn't exist, then the DNA would not amplify and a negative result would occur. The DNA sequence for the Single Nucleotide Polymorphism is shown below, it is a mutation in the CHEK2 gene which is found on chr22:29083730-29137821 or Chromosome 22- base pairs 29083730 to 29137821. The mutation of the gene is the addition of a Cytosine rather than a Thymine. This is shown in the SNP below. Primer Development: With the DNA sequence (SNP) below: 5' GGAAGTGGGTCCTAAAAACTCTTACA[C/T]TGCATACATAGAAGATCACAGTGGC 3' The forward primer would be: 3' CAGGATTTTTGTGAATGTGAG 5'
3' CACTGCATACATAGAAGATCA 5' This is within the accepted bp primer length (18-22), follows the GC concentration rule (40-60%), follows the GC clamp rule (G or C within 5 bp of 3' to clamp the primer down), and has an annealing temperature of 61 degrees Celsius forward and 59 degrees Celsius backward. These all show that the primers forward and backward for this strand above would work. The primer also contains the mutation from the DNA sequence. Bayes Analysis Bayes analysis accounts for probabilities behind a test. In this example, Bayes rule is use to calculate the probability that a test will give a positive result for cancer when cancer is present: [math]\displaystyle{ p(C|T)= p(T|C)*p(C) / p(T|C)*p(C)+p(T|nC)*p(nC) }[/math] This is where: p(C|T)= probability that cancer present when positive test p(T|C)= probability that positive test when cancer present p(C)= probability of cancer p(T|nC)= probability that positive test when cancer not present p(nC)= probability that cancer not present Meijers-Heijboer et al. (2002) showed that having the CHEK2 gene creates a higher risk, but not always cancer. 1.1% have the gene, but do not have the cancer. 5.1% have the gene and have breast cancer. This was done from 718 families that did not have BRCA1 or BRCA2 mutations. This shows that the cancer gene could be present but still would not mean cancer is present. Dong et al. (2003) measured that 28 out of 578 men (4.8%) have a CHEK2 gene mutation and have prostate cancer. Cybulski et al. (2004) found that in Poland, variants of the CHEK2 gene have a strong link to increase risk of prostate cancer. In Poland, the prevalence of the alleles in 4008 cancer cases and 4000 controls shows the importance in the link between the CHEK2 gene variants and prostate cancer. These variables are found from the OMIM entry Checkpoint Kinase 2. Other factors would be inherent in experimental errors. However, the limited amount of data found through PCR done in this experiment does not allow for a calculated percentage.
All images borrowed from Virtual Lab via the University of Utah.
Results
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||