BME103:T930 Group 3
| Home People Lab Write-Up 1 Lab Write-Up 2 Lab Write-Up 3 Course Logistics For Instructors Photos Wiki Editing Help | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
OUR TEAM
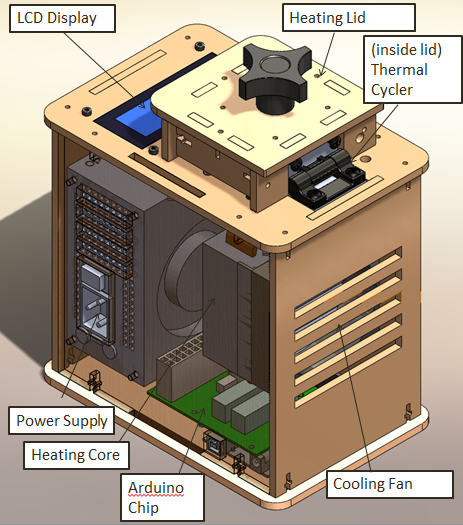
LAB 1 WRITE-UPInitial Machine TestingThe Original Design
When we unplugged the LCD display wire(part 3) from the Arudnio chip(part 6), the screen turned off. Everything on the PCR was working fine expect there was no output on the display. When we unplugged the white wire that connects Arudino chip(part 6) to thermal cycler (part 2), the reading from the screen dropped to -40 degrees Celsius. We disconnected the wire multiple times and each time the screen displayed -40 degrees Celsius.
We ran a test run on 10/25/2012. For this test we placed some empty PCR tubes into the machine and ran a simple test program on the Open PCR software. After the simple test was over we noticed that the display screen on the Open PCR lid matched very closely with what was displayed on our computer screen. The agreement between our computer screen and our PCR display meant that our diagnostic test was a success.
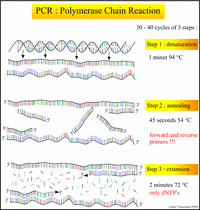
ProtocolsPolymerase Chain Reaction Polymerase Chain Reaction Procedure: 2.)We labeled 8 empty PCR tubes. For the first sample we labeled the 3 DNA samples 1A, 1B and 1C. For the second sample we labeled the tubes 2A, 2B and 2C. For the positive and negative controls, we labeled the tubes + and - respectively. 3.)Using one pipette per sample, to avoid contamination, we transferred the PCR reaction mix we were given to the PCR tubes. 4.)We then placed the samples in the PCR machine 5.)We set our PCR program to three stages. Stage one: 1 cycle, 95 degree Celsius for 3 minutes. Stage 2: 35 cycles, 95 degrees Celsius for 30 seconds, 57 degrees Celsius for 30 seconds, 72 degrees Celsius. Stage three: 72 degrees Celsius for 3 minutes and then hold at 4 degree Celsius.
Fluorimeter Assembly
Fluorimeter Procedure: 2.)With the permanent marker we also labeled the Eppendrof tubes at the top, we had a total of 10 Eppendrof tubes labeled and 10 pipettes labeled. 3.)We transferred each sample separately into the Eppendorf tubes containing 400 ml of buffer. 4.)Using a specially labeled Eppendorf tube containing SYBR GREEN, with its own pippter, we placed two drops onto the first two center drops. 5.)Then using the sample we placed two drops on top of the SYBR GREEN solution drops 6.)Then we aligned the blue light to pass through the drop. 7.)Then the smartphone operator took a picture with the settings on the phone adjusted to inactive flash, iso to 800, white balance to auto, exposure to the highest setting and contrast to the lowest setting. 8.)This process was repeated for all samples 9.)After picture was taken it was given to the Image J software
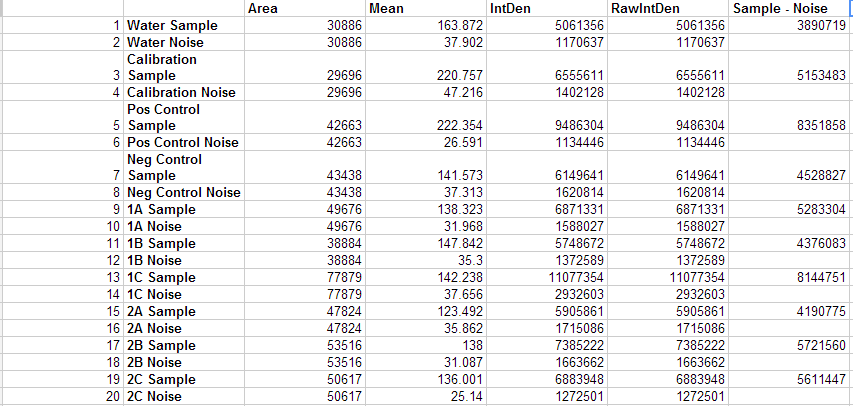
Image J Procedure: 2.) Using the menu selection we used, image > color > split channels 3.) This created 3 files: a red, green and blue image. We only used the green image. 4.) Then we clicked the menu bar to activate the oval selection. 5.) We drew an oval around the image of our sample and then selected analyze > measure. 6.) We then simply moved the previously drawn oval to an area away from the sample to obtain the noise measurements. 7.) We repeated this process for all of the samples, including the controls.
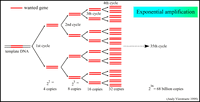
Research and DevelopmentSpecific Cancer Marker Detection - The Underlying Technology In order to isolate and detect the cancer-causing gene, we essentially added a very specific primer that attaches to the cancer gene on the DNA. This Primer will not only allow for replication of the DNA, but more specifically, replication of the cancer gene in the DNA sample. The Open PCR machine then manipulates the temperature of the sample to facilitate the constant replication of the isolated DNA strands. We cycled the machine 30 times (the DNA was replicated 30 times) to be sure we had enough of the cancer DNA present in our solution. By understanding the mechanism of the Open PCR, we can intuitively understand how genes with specific diseases - such as cancer - are amplified. It is now necessary to learn how to identify which samples contain those replicated genes. The rs17879961 cancer-associated sequence in particular, can be identified within the DNA because of the single nucleotide variation - in this case a missense mutation - in its gene code. The mutation will yield a positive identification marker for cancer when the nucleotide cytosine is replaced by thymine in a very specific section of the DNA. Original Code: Code with SNP: Our research of the DNA sequence rs17879961 reveals that the gene sequence is correlated most directly to Breast and Colorectal Cancer. The Protocol section explains how the Open PCR amplifies this specific DNA sequence. A non-cancer DNA sequence would not produce a signal because the primer would not bind to the DNA and therefore would not allow for any replication. Only when we have the specific missense mutation can we replicate the cancer gene and identify a positive signal from the DNA. Although we determined that the DNA sequence rs17879961 was correlated to the Breast and Colorectal cancer, we must look at Baye's rule and other statistics to understand the likelihood that the gene will in fact lead to the development and spread of the cancer. Bayesian reasoning accounts for the prevalence, sensitivity, specificity and false positive rates of a disease in order to further understand that disease. Sensitivity is the likelihood that a person afflicted with the disease will test positive for it, while specificity is the likelihood that a person without the disease will test negative for it. Conditional probabilities of a sample of 180 people in Finland determined that the frequency of this cancer found in Finland was 7.8%. The genotype of this single nucleotide polymorphism change from cytosine to thymine in this population was 1.1% and the genotype without the SNP was found to be 98.9%. For more infomation, visit http://www.ncbi.nlm.nih.gov/projects/SNP/snp_ref.cgi?rs=17879961. BONUS points: Use a program like Powerpoint, Word, Illustrator, Microsoft Paint, etc. to illustrate how primers bind to the cancer DNA template, and how Taq polymerases amplify the DNA. Screen-captures from the OpenPCR tutorial might be useful. Be sure to credit the source if you borrow images. 1.) Both of the above images: http://users.ugent.be/~avierstr/principles/pcr.html
Results
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||